
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,893,012 – 17,893,150 |
| Length | 138 |
| Max. P | 0.957439 |

| Location | 17,893,012 – 17,893,130 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -24.30 |
| Energy contribution | -24.93 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17893012 118 + 22224390 CAUAUGUGUUUAUAUAUUAUAUUAUUCCCAGUGUAGCGCCACAUGCAAGAUGUUGUCCCCUUUUAUUUAUUUGUGGAAAAGGGCAUCCCGGGGGCAGUGGCUAAACUGGACACAAAAU ....(((((....(((......)))..(((((.....(((((..((.....))(((((((((((((......))))))..(((...)))))))))))))))...)))))))))).... ( -34.60) >DroSec_CAF1 157070 111 + 1 CAUAUGU------AUAUAAUAUUAUUCCCAGUGUAGCGCCACAUGCAAGAUGUUGUCC-CUUUUAUUUAUUUGUGGAAAAGGGCAUCCCGGGAGCAGUGGCUAAACUGGACACAAAAU .......------..............(((((.....(((((.(((.......((.((-(((((............)))))))))((....))))))))))...)))))......... ( -27.90) >DroEre_CAF1 152526 108 + 1 ----UA-----AUGUAUAAUAUUAUUCCCAGUGUAGCGCCACAUGCAAGAUGUCGUCC-CUUUAAUUUAUUUGGGAAAAGGGGCAUCCCGGGGGCAGUGGCUAAACUGGACACAAAAU ----..-----................(((((.....(((((.(((..((((((.(((-(............)))).....))))))(....)))))))))...)))))......... ( -30.30) >DroYak_CAF1 153410 107 + 1 ----UA------AAUACGACAUUAUUCCCAGUGUAGUGCCACAUGCAAGAUGUUGUCC-CUUUUAUUUAUUGGGCGAAAGGGGCAUCCCGGGGGCAGUGGCUAAACUGGACACAAAAU ----..------...............(((((.....(((((.(((.......(((((-((((..((.....))..)))))))))........))))))))...)))))......... ( -33.76) >consensus ____UA______UAUAUAAUAUUAUUCCCAGUGUAGCGCCACAUGCAAGAUGUUGUCC_CUUUUAUUUAUUUGGGGAAAAGGGCAUCCCGGGGGCAGUGGCUAAACUGGACACAAAAU ...........................(((((.....(((((.(((........((((..((((((......))))))..)))).((....))))))))))...)))))......... (-24.30 = -24.93 + 0.63)


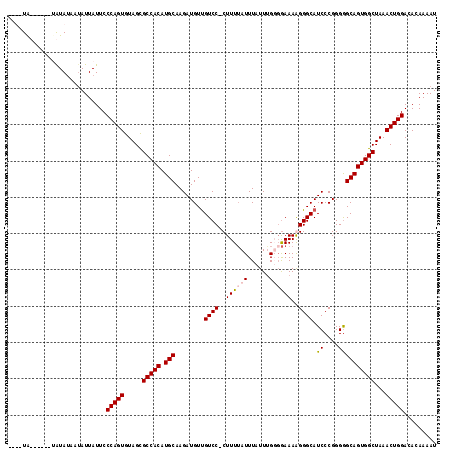
| Location | 17,893,012 – 17,893,130 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -24.80 |
| Energy contribution | -24.55 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17893012 118 - 22224390 AUUUUGUGUCCAGUUUAGCCACUGCCCCCGGGAUGCCCUUUUCCACAAAUAAAUAAAAGGGGACAACAUCUUGCAUGUGGCGCUACACUGGGAAUAAUAUAAUAUAUAAACACAUAUG ....((((((((((...((((((((....(((((((((((((.............)))))))....))))))))).))))).....)))))......((((...)))).))))).... ( -30.32) >DroSec_CAF1 157070 111 - 1 AUUUUGUGUCCAGUUUAGCCACUGCUCCCGGGAUGCCCUUUUCCACAAAUAAAUAAAAG-GGACAACAUCUUGCAUGUGGCGCUACACUGGGAAUAAUAUUAUAU------ACAUAUG ...((((..(((((...((((((((....(((((((((((((............)))))-))....))))))))).))))).....)))))..))))........------....... ( -30.30) >DroEre_CAF1 152526 108 - 1 AUUUUGUGUCCAGUUUAGCCACUGCCCCCGGGAUGCCCCUUUUCCCAAAUAAAUUAAAG-GGACGACAUCUUGCAUGUGGCGCUACACUGGGAAUAAUAUUAUACAU-----UA---- ...((((..(((((...((((((((....(((((((((((((.............))))-))..).))))))))).))))).....)))))..))))..........-----..---- ( -30.52) >DroYak_CAF1 153410 107 - 1 AUUUUGUGUCCAGUUUAGCCACUGCCCCCGGGAUGCCCCUUUCGCCCAAUAAAUAAAAG-GGACAACAUCUUGCAUGUGGCACUACACUGGGAAUAAUGUCGUAUU------UA---- ...((((..(((((.((((((((((....((((((.((((((.............))))-))....))))))))).)))))..)).)))))..)))).........------..---- ( -29.02) >consensus AUUUUGUGUCCAGUUUAGCCACUGCCCCCGGGAUGCCCCUUUCCACAAAUAAAUAAAAG_GGACAACAUCUUGCAUGUGGCGCUACACUGGGAAUAAUAUUAUAUA______CA____ ...((((..(((((...((((((((....((((((.((((((.............)))).))....))))))))).))))).....)))))..))))..................... (-24.80 = -24.55 + -0.25)


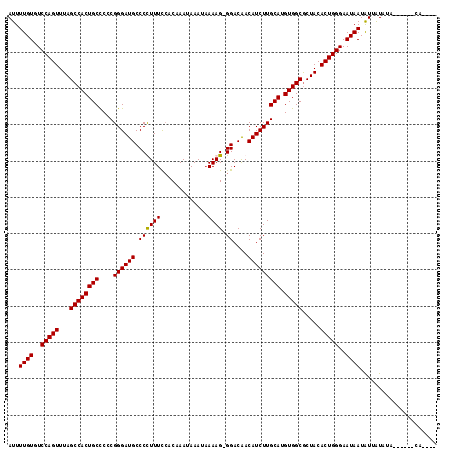
| Location | 17,893,051 – 17,893,150 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 94.59 |
| Mean single sequence MFE | -25.49 |
| Consensus MFE | -20.37 |
| Energy contribution | -20.12 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17893051 99 - 22224390 CAGCUAAAGUGCUCAUAAACAUUUUGUGUCCAGUUUAGCCACUGCCCCCGGGAUGCCCUUUUCCACAAAUAAAUAAAAGGGGACAACAUCUUGCAUGUG ..((((((.((..(((((.....)))))..)).))))))((((((....(((((((((((((.............)))))))....))))))))).))) ( -25.42) >DroSec_CAF1 157103 98 - 1 CAGCUAAAGUGCUCAUAAACAUUUUGUGUCCAGUUUAGCCACUGCUCCCGGGAUGCCCUUUUCCACAAAUAAAUAAAAG-GGACAACAUCUUGCAUGUG ..((((((.((..(((((.....)))))..)).))))))((((((....(((((((((((((............)))))-))....))))))))).))) ( -25.60) >DroEre_CAF1 152556 98 - 1 CAGCUAAAGUGCUCAUAAAGAUUUUGUGUCCAGUUUAGCCACUGCCCCCGGGAUGCCCCUUUUCCCAAAUAAAUUAAAG-GGACGACAUCUUGCAUGUG ..((((((.((..(((((.....)))))..)).))))))((((((....(((((((((((((.............))))-))..).))))))))).))) ( -25.82) >DroYak_CAF1 153439 98 - 1 CAGCUAAAGUGCUCAUAAACAUUUUGUGUCCAGUUUAGCCACUGCCCCCGGGAUGCCCCUUUCGCCCAAUAAAUAAAAG-GGACAACAUCUUGCAUGUG ..((((((.((..(((((.....)))))..)).))))))((((((....((((((.((((((.............))))-))....))))))))).))) ( -25.12) >consensus CAGCUAAAGUGCUCAUAAACAUUUUGUGUCCAGUUUAGCCACUGCCCCCGGGAUGCCCCUUUCCACAAAUAAAUAAAAG_GGACAACAUCUUGCAUGUG ..((((((.((..(((((.....)))))..)).))))))((((((....((((((.((((((.............)))).))....))))))))).))) (-20.37 = -20.12 + -0.25)

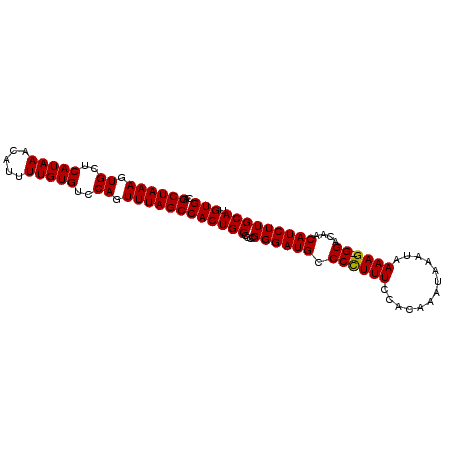
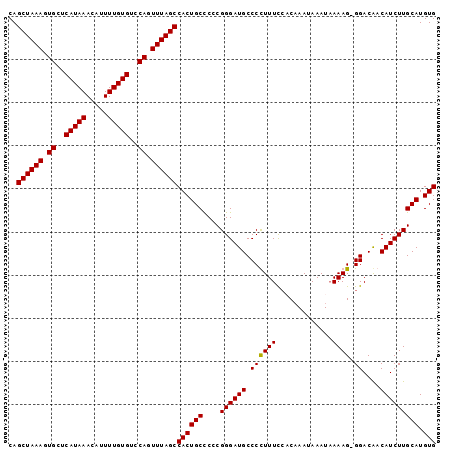
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:55 2006