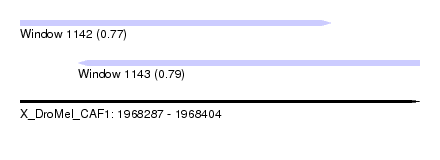
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,968,287 – 1,968,404 |
| Length | 117 |
| Max. P | 0.791249 |

| Location | 1,968,287 – 1,968,378 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 95.65 |
| Mean single sequence MFE | -24.42 |
| Consensus MFE | -20.88 |
| Energy contribution | -21.28 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
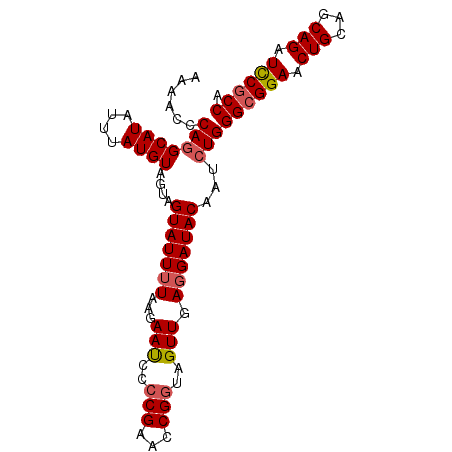
>X_DroMel_CAF1 1968287 91 + 22224390 AAAACCCA-GCAUAUUUAUGUAGUAGUAUUGUAAGAAUCCCCGAACCGGUAGUUGAGGAUACAAUCUGGGCGGAACUGCAGCAGAUCCGCCA ........-((((....))))..(((.((((((....(((.(((........))).))))))))))))((((((.(((...))).)))))). ( -23.00) >DroSec_CAF1 1173 92 + 1 AAAACCCAGGCAUAUUUAUGUAGUAGUAUUUUAAGAAUCCCCGAACCGAUAGUUGAGGAUACAAUCUGGGCGGAACUGCAGCAGAUCCGCCA ......(((((((....)))).............(.((((.(((........))).)))).)...)))((((((.(((...))).)))))). ( -22.40) >DroSim_CAF1 1171 92 + 1 AAAACCCAGGCAUAUUUAUGUAGUAGUAUUUUAAGAAUCCCCGAACCGGUAGUUGAGGAUACAAUCUGGGCGGAACUGCAGCAGAUCCGCCA ......(((((((....))))....(((((((...(((..(((...)))..))).)))))))...)))((((((.(((...))).)))))). ( -25.50) >DroEre_CAF1 1209 92 + 1 AAAACCCAGGCAUAUUUAUGUAGUAGUAUUUUAAGAACCCCCGAACCGGAAGUUGAGGAUACAAUCUGGGCGGAACUGCAGCAGAUCCGCCA ......(((((((....))))....(((((((...(((..(((...)))..))).)))))))...)))((((((.(((...))).)))))). ( -25.70) >DroYak_CAF1 1234 92 + 1 AAAACCCAGGCAUAUUUAUGUAGUAGUAUUUUAAGAACCUCCGAACCGGAAGUUGAGGAUACAAUCUGGGCGGAACUGCAGCAGUUUCCCCA ....(((((((((....))))....(((((((...(((.((((...)))).))).)))))))...))))).(((((((...))))))).... ( -25.50) >consensus AAAACCCAGGCAUAUUUAUGUAGUAGUAUUUUAAGAAUCCCCGAACCGGUAGUUGAGGAUACAAUCUGGGCGGAACUGCAGCAGAUCCGCCA ......(((((((....))))....(((((((...(((..(((...)))..))).)))))))...)))((((((.(((...))).)))))). (-20.88 = -21.28 + 0.40)

| Location | 1,968,304 – 1,968,404 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -19.22 |
| Energy contribution | -20.38 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
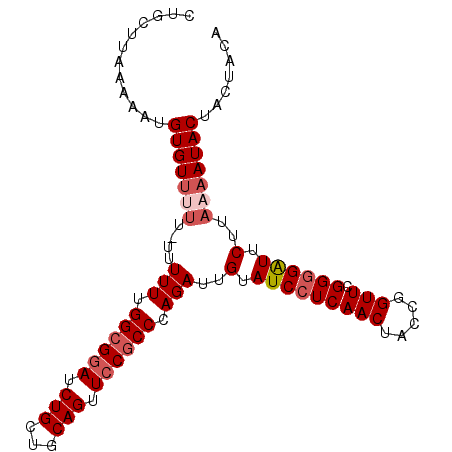
>X_DroMel_CAF1 1968304 100 - 22224390 CUGCUUAAAAAUGUGUUUUUGUUUUUUGGCGGAUCUGCUGCAGUUCCGCCCAGAUUGUAUCCUCAACUACCGGUUCGGGGAUUCUUACAAUACUACUACA (((...(((((((......))))))).((((((.(((...))).)))))))))((((((((((((((.....))).)))))....))))))......... ( -25.20) >DroSec_CAF1 1191 98 - 1 CUGCUUAAAAAUGUGUUUUU--UUUUUGGCGGAUCUGCUGCAGUUCCGCCCAGAUUGUAUCCUCAACUAUCGGUUCGGGGAUUCUUAAAAUACUACUACA ............(((((((.--.(((.((((((.(((...))).)))))).)))..(.(((((((((.....))).)))))).)..)))))))....... ( -25.20) >DroSim_CAF1 1189 97 - 1 CUGCUUAAAAAUGUGUUUUU---UUUUGGCGGAUCUGCUGCAGUUCCGCCCAGAUUGUAUCCUCAACUACCGGUUCGGGGAUUCUUAAAAUACUACUACA ............(((((((.---(((.((((((.(((...))).)))))).)))..(.(((((((((.....))).)))))).)..)))))))....... ( -25.20) >DroEre_CAF1 1227 100 - 1 CUUCUCAAACAUGUGUGUUUCUUUUUUGGCGGAUCUGCUGCAGUUCCGCCCAGAUUGUAUCCUCAACUUCCGGUUCGGGGGUUCUUAAAAUACUACUACA ............((((((((...(((.((((((.(((...))).)))))).)))..(.(((((((((.....))).)))))).)...)))))).)).... ( -24.30) >DroYak_CAF1 1252 97 - 1 CAUCUUAAAAAUGUGUU---UUUUUUUGGGGAAACUGCUGCAGUUCCGCCCAGAUUGUAUCCUCAACUUCCGGUUCGGAGGUUCUUAAAAUACUACUACA ............(((((---((..(((((((.(((((...))))).).))))))..........((((((((...))))))))...)))))))....... ( -22.70) >consensus CUGCUUAAAAAUGUGUUUUU_UUUUUUGGCGGAUCUGCUGCAGUUCCGCCCAGAUUGUAUCCUCAACUACCGGUUCGGGGAUUCUUAAAAUACUACUACA ............(((((((....(((.((((((.(((...))).)))))).)))..(.(((((((((.....))).)))))).)..)))))))....... (-19.22 = -20.38 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:47 2006