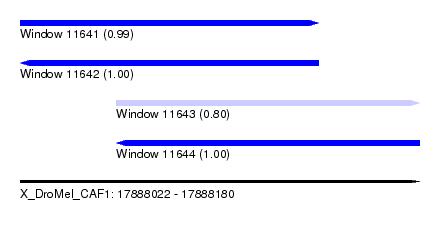
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,888,022 – 17,888,180 |
| Length | 158 |
| Max. P | 0.998077 |

| Location | 17,888,022 – 17,888,140 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.77 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -18.16 |
| Energy contribution | -18.44 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17888022 118 + 22224390 --UUAAUUUAAAGCAACCGCUUUUAAUAAUCACCAUUUAUUGCAACUCGCAAACUGCUCUCAGCAAAUUCAUAUGAAUUCCAAUUACCAAUUAUGGGAAAAAGCAGUUUGCUUACUUUUG --...(((.(((((....))))).))).....................((((((((((.((....(((((....))))).......(((....)))))...))))))))))......... ( -23.30) >DroSec_CAF1 151999 118 + 1 --UUAAUUUAAAGCAACCGCUUUUAAUAAUCACCAUUUAUUGCAACUCGCAAACUGCUCUCAGCAAAUUCAUAUAAAUUCCAAUUACCAAUUAUGGGAAAAAGCAGUUUGCUUACUUCUG --...(((.(((((....))))).))).....................((((((((((((((...............................)))))....)))))))))......... ( -19.88) >DroSim_CAF1 130705 118 + 1 --UUAAUUUAAAGCAACCGCUUUUAAUAAUCACCAUUUAUUGCAACUCGCAAACUGCUCUCAGCGAAUUCAUAUGAAUUCCAAUUACCAAUUAUGGGAAAAAGCAGUUUGCUUACUUCUG --...(((.(((((....))))).))).....................((((((((((.((.(.((((((....))))))).....(((....)))))...))))))))))......... ( -27.20) >DroEre_CAF1 147864 120 + 1 UAUUAAUUUAAAGCACACGCUUUUAAUAAUCACCAUUUAUUGCAACUUGCGAAAUGCUCUCAGGAAAUUUACAUGAAUUUCAAUUACCAAUUAUGGGAAAAAGCAGUUUGCUUACUUCUG ..........(((((.((((((((.........((((..((((.....)))))))).(((((.(((((((....)))))))............))))))))))).)).)))))....... ( -22.12) >DroYak_CAF1 148370 118 + 1 --AUAAUUUAAAGCACCCGCUUUUAAUAAUCACCAUUUAUUGCAACUUGCAAAAUGCUCUCAGCAAAUUUACAUGAAUUCCAAUUAACAAUUAUGGGAAAAAGCAGUUUGCUUACUUCUG --........(((((..(((((((.....(((.......((((.....))))..(((.....)))........))).(((((...........))))))))))).)..)))))....... ( -19.80) >consensus __UUAAUUUAAAGCAACCGCUUUUAAUAAUCACCAUUUAUUGCAACUCGCAAACUGCUCUCAGCAAAUUCAUAUGAAUUCCAAUUACCAAUUAUGGGAAAAAGCAGUUUGCUUACUUCUG .....(((.(((((....))))).))).....................((((((((((.(((...........))).(((((...........)))))...))))))))))......... (-18.16 = -18.44 + 0.28)


| Location | 17,888,022 – 17,888,140 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.77 |
| Mean single sequence MFE | -29.31 |
| Consensus MFE | -25.26 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17888022 118 - 22224390 CAAAAGUAAGCAAACUGCUUUUUCCCAUAAUUGGUAAUUGGAAUUCAUAUGAAUUUGCUGAGAGCAGUUUGCGAGUUGCAAUAAAUGGUGAUUAUUAAAAGCGGUUGCUUUAAAUUAA-- ..((((((((((((((((((((..(((....)))......((((((....))))))...))))))))))))).(((..(........)..)))...........))))))).......-- ( -31.70) >DroSec_CAF1 151999 118 - 1 CAGAAGUAAGCAAACUGCUUUUUCCCAUAAUUGGUAAUUGGAAUUUAUAUGAAUUUGCUGAGAGCAGUUUGCGAGUUGCAAUAAAUGGUGAUUAUUAAAAGCGGUUGCUUUAAAUUAA-- ..((((((((((((((((((((..(((....)))......((((((....))))))...))))))))))))).(((..(........)..)))...........))))))).......-- ( -29.70) >DroSim_CAF1 130705 118 - 1 CAGAAGUAAGCAAACUGCUUUUUCCCAUAAUUGGUAAUUGGAAUUCAUAUGAAUUCGCUGAGAGCAGUUUGCGAGUUGCAAUAAAUGGUGAUUAUUAAAAGCGGUUGCUUUAAAUUAA-- ..((((((((((((((((((((..(((....)))......((((((....))))))...))))))))))))).(((..(........)..)))...........))))))).......-- ( -34.30) >DroEre_CAF1 147864 120 - 1 CAGAAGUAAGCAAACUGCUUUUUCCCAUAAUUGGUAAUUGAAAUUCAUGUAAAUUUCCUGAGAGCAUUUCGCAAGUUGCAAUAAAUGGUGAUUAUUAAAAGCGUGUGCUUUAAAUUAAUA .......(((((.((.((((((((((((.....((((((((((((......))))))......((.....)).)))))).....)))).)).....)))))))).))))).......... ( -22.20) >DroYak_CAF1 148370 118 - 1 CAGAAGUAAGCAAACUGCUUUUUCCCAUAAUUGUUAAUUGGAAUUCAUGUAAAUUUGCUGAGAGCAUUUUGCAAGUUGCAAUAAAUGGUGAUUAUUAAAAGCGGGUGCUUUAAAUUAU-- .......(((((..((((((((.......((..((.((((.((((..((((((..(((.....))).)))))))))).)))).))..)).......)))))))).)))))........-- ( -28.64) >consensus CAGAAGUAAGCAAACUGCUUUUUCCCAUAAUUGGUAAUUGGAAUUCAUAUGAAUUUGCUGAGAGCAGUUUGCGAGUUGCAAUAAAUGGUGAUUAUUAAAAGCGGUUGCUUUAAAUUAA__ ..((((((.(((((((((((((..(((....)))......((((((....))))))...))))))))))))).(((..(........)..)))............))))))......... (-25.26 = -25.90 + 0.64)



| Location | 17,888,060 – 17,888,180 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -20.66 |
| Energy contribution | -21.94 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17888060 120 + 22224390 UGCAACUCGCAAACUGCUCUCAGCAAAUUCAUAUGAAUUCCAAUUACCAAUUAUGGGAAAAAGCAGUUUGCUUACUUUUGGCAAAAUUGUUGCUAAAAUGUGUAAAAACAAACUGUUGUG .(((((..((((((((((.((....(((((....))))).......(((....)))))...)))))))))).((((((((((((.....))))))))).)))............))))). ( -32.80) >DroSec_CAF1 152037 120 + 1 UGCAACUCGCAAACUGCUCUCAGCAAAUUCAUAUAAAUUCCAAUUACCAAUUAUGGGAAAAAGCAGUUUGCUUACUUCUGGCCAAAUUGUUGCUAAAAUGUGUAAAACCAAACUGUUGUG .(((((..((((((((((((((...............................)))))....))))))))).(((((.((((.........)))).)).)))............))))). ( -22.58) >DroSim_CAF1 130743 120 + 1 UGCAACUCGCAAACUGCUCUCAGCGAAUUCAUAUGAAUUCCAAUUACCAAUUAUGGGAAAAAGCAGUUUGCUUACUUCUGGCCAAAUUGUUGCUAAAAUGUGUAAAACCAAACUGUGUUG ..((((.(((((((((((.((.(.((((((....))))))).....(((....)))))...)))))))))).(((((.((((.........)))).)).)))............).)))) ( -28.00) >DroEre_CAF1 147904 120 + 1 UGCAACUUGCGAAAUGCUCUCAGGAAAUUUACAUGAAUUUCAAUUACCAAUUAUGGGAAAAAGCAGUUUGCUUACUUCUGGCAAAAAUGUUGCUAAAAUGUGUAAAAACAAACUGUUGUG .(((((..(((((.((((.((..(((((((....))))))).....(((....)))))...)))).))))).(((((.((((((.....)))))).)).)))............))))). ( -25.40) >DroYak_CAF1 148408 120 + 1 UGCAACUUGCAAAAUGCUCUCAGCAAAUUUACAUGAAUUCCAAUUAACAAUUAUGGGAAAAAGCAGUUUGCUUACUUCUGGCAAAAUUGUUGCUUAAAUGUGUAAAAACAAACUGUUGUG .(((((........(((.....)))..(((((((...(((((...........)))))..((((((((((((.......))))))....))))))....)))))))........))))). ( -23.90) >consensus UGCAACUCGCAAACUGCUCUCAGCAAAUUCAUAUGAAUUCCAAUUACCAAUUAUGGGAAAAAGCAGUUUGCUUACUUCUGGCAAAAUUGUUGCUAAAAUGUGUAAAAACAAACUGUUGUG .(((((..((((((((((.(((...........))).(((((...........)))))...)))))))))).(((((.((((((.....)))))).)).)))............))))). (-20.66 = -21.94 + 1.28)

| Location | 17,888,060 – 17,888,180 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -28.76 |
| Consensus MFE | -25.66 |
| Energy contribution | -26.86 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17888060 120 - 22224390 CACAACAGUUUGUUUUUACACAUUUUAGCAACAAUUUUGCCAAAAGUAAGCAAACUGCUUUUUCCCAUAAUUGGUAAUUGGAAUUCAUAUGAAUUUGCUGAGAGCAGUUUGCGAGUUGCA ...........................(((((....((((.....))))(((((((((((((..(((....)))......((((((....))))))...)))))))))))))..))))). ( -32.80) >DroSec_CAF1 152037 120 - 1 CACAACAGUUUGGUUUUACACAUUUUAGCAACAAUUUGGCCAGAAGUAAGCAAACUGCUUUUUCCCAUAAUUGGUAAUUGGAAUUUAUAUGAAUUUGCUGAGAGCAGUUUGCGAGUUGCA ..((((..(((((((......................))))))).....(((((((((((((..(((....)))......((((((....))))))...)))))))))))))..)))).. ( -29.95) >DroSim_CAF1 130743 120 - 1 CAACACAGUUUGGUUUUACACAUUUUAGCAACAAUUUGGCCAGAAGUAAGCAAACUGCUUUUUCCCAUAAUUGGUAAUUGGAAUUCAUAUGAAUUCGCUGAGAGCAGUUUGCGAGUUGCA ((((....(((((((......................))))))).....(((((((((((((..(((....)))......((((((....))))))...)))))))))))))..)))).. ( -34.05) >DroEre_CAF1 147904 120 - 1 CACAACAGUUUGUUUUUACACAUUUUAGCAACAUUUUUGCCAGAAGUAAGCAAACUGCUUUUUCCCAUAAUUGGUAAUUGAAAUUCAUGUAAAUUUCCUGAGAGCAUUUCGCAAGUUGCA ..(((((((((((((....((.((((.((((.....)))).)))))))))))))))((((((..(((....))).....((((((......))))))..)))))).........)))).. ( -23.90) >DroYak_CAF1 148408 120 - 1 CACAACAGUUUGUUUUUACACAUUUAAGCAACAAUUUUGCCAGAAGUAAGCAAACUGCUUUUUCCCAUAAUUGUUAAUUGGAAUUCAUGUAAAUUUGCUGAGAGCAUUUUGCAAGUUGCA ..((((................(((((..(((((((.((...((((.((((.....)))))))).)).)))))))..))))).....((((((..(((.....))).)))))).)))).. ( -23.10) >consensus CACAACAGUUUGUUUUUACACAUUUUAGCAACAAUUUUGCCAGAAGUAAGCAAACUGCUUUUUCCCAUAAUUGGUAAUUGGAAUUCAUAUGAAUUUGCUGAGAGCAGUUUGCGAGUUGCA ...........................(((((....((((.....))))(((((((((((((..(((....)))......((((((....))))))...)))))))))))))..))))). (-25.66 = -26.86 + 1.20)

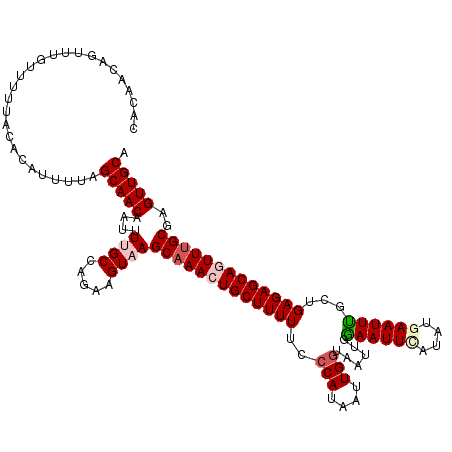

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:52 2006