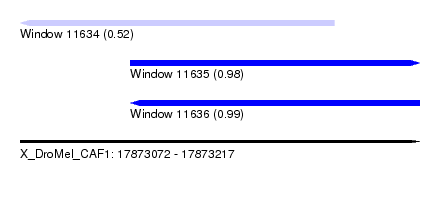
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,873,072 – 17,873,217 |
| Length | 145 |
| Max. P | 0.986228 |

| Location | 17,873,072 – 17,873,186 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.13 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -31.69 |
| Energy contribution | -31.58 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17873072 114 - 22224390 UU------GGGAAACAGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAGCUGAUGGGCAUCGUCCACAAGGAGCGGCGCCA ..------.((...(.(((((((...((.((....))))...))))))).((((((.(....)))))))......(((.(((((((.(((....))).)))))))...)))..)...)). ( -47.70) >DroVir_CAF1 188395 105 - 1 ---------------AGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAACUGAUGGGAAUCGUGCACAAGGAGCGCCGACA ---------------.(((((((...((.((....))))...)))))))...(..(((.(((..(((((.....(((....)))........)))))..)))((.......)))))..). ( -39.62) >DroGri_CAF1 163724 105 - 1 ---------------AGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAACUGAUGGGCAUCGUGCACAAGGAGCGCCGCCA ---------------.(((((((...((.((....))))...)))))))...((.(((....((((((((....(((....)))....))).((.((.....)).)).)))))))).)). ( -42.00) >DroWil_CAF1 164536 120 - 1 UAAAGGGUAAAAAAAAGCCGCCAAGAUGACCAUGUGGCAGAGUGGUGGUAUGGGAGGCCAUGGCUCCCAAAAUAAUCCAUGGAUGAAACUAAUGGGCAUUGUCCAUAAGGAGCGGCGACA ....((((........(((((((...((.((....))))...))))))).((((((.(....))))))).....)))).....((...((.((((((...)))))).))...))...... ( -37.70) >DroMoj_CAF1 207614 111 - 1 ---------CGCACCAAACGCCAAGAUGACGAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAGCUGAUGGGCAUCGUGCACAAGGAGCGACGGCA ---------....(((..(((((...((.(......)))...)))))...)))...(((.(.((((((((....(((....)))....))).((.((.....)).)).))))).).))). ( -36.60) >DroAna_CAF1 145739 117 - 1 GU---GACGACCAACAGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAGCUGAUGGGCAUUGUCCACAAGGAGCGACGCCA ..---.....(((...(((((((...((.((....))))...))))))).)))..(((..(.((((((((....(((....)))....))).(((((...)))))...))))).).))). ( -45.50) >consensus ____________AACAGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAUGGAUGAAACUGAUGGGCAUCGUCCACAAGGAGCGACGCCA ................(((((((...((.((....))))...))))))).((((((.(....)))))))......(((.(((((((..((....))..)))).).)).)))......... (-31.69 = -31.58 + -0.11)

| Location | 17,873,112 – 17,873,217 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.72 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -24.35 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17873112 105 + 22224390 AUGGAUUGUUCUGGGAGCCAUGGCCUCCCAUGCCGCCACUCUGCCACAUGGUCAUCUUGGCGGCUGUUUCCC------AAAUCC---GGUUCCGAUCGAAGAAACUG------UGGCAGU .(((...((..(((((((....).)))))).))..)))..((((((((...((.((((((.(((((..(...------..)..)---))))))))..)).))...))------)))))). ( -35.00) >DroPse_CAF1 144879 90 + 1 AUGGAUUGUUCUGGGAGCCAUGGCCUCCCAUGCCGCCACUCUGCCACAUAGUCAUCUUGGCGGCUCUCUUUG------CACUCU---G---------------ACUG------UGGCAGU .(((...((..(((((((....).)))))).))..)))..((((((((..((((.....((((......)))------)....)---)---------------))))------)))))). ( -32.90) >DroWil_CAF1 164576 100 + 1 AUGGAUUAUUUUGGGAGCCAUGGCCUCCCAUACCACCACUCUGCCACAUGGUCAUCUUGGCGGCUUUUUUUUACCCUUUAAUCC---UUUU-----------AAAAG------UGGCAGU .(((.......(((((((....).)))))).....)))..(((((((...(((.....)))...............(((((...---..))-----------))).)------)))))). ( -27.10) >DroYak_CAF1 132415 111 + 1 AUGGAUUGUUCUGGGAGCCAUGGCCUCCCAUGCCGCCACUCUGCCACAUGGUCAUCUUGGCGGCUGUUUCCC------AAAUCC---GUUUCCGAUCGAAGAAACUGUGUCUGUGGCAGU ..(((((....(((((((....).)))))).(((((((...((((....)).))...)))))))........------.)))))---(((((........)))))..(((.....))).. ( -37.00) >DroAna_CAF1 145779 96 + 1 AUGGAUUGUUCUGGGAGCCAUGGCCUCCCAUGCCGCCACUCUGCCACAUGGUCAUCUUGGCGGCUGUUGGUCGUC---ACCUCCUUGGCCUCGGAUCGA--------------------- .....(((.((((((.((((((((..((...(((((((...((((....)).))...)))))))....))..)))---)......)))))))))).)))--------------------- ( -37.10) >DroPer_CAF1 148743 90 + 1 AUGGAUUGUUCUGGGAGCCAUGGCCUCCCAUGCCGCCACUCUGCCACAUAGUCAUCUUGGCGGCUCUCUUUG------CACUCU---G---------------ACUG------UGGCAGU .(((...((..(((((((....).)))))).))..)))..((((((((..((((.....((((......)))------)....)---)---------------))))------)))))). ( -32.90) >consensus AUGGAUUGUUCUGGGAGCCAUGGCCUCCCAUGCCGCCACUCUGCCACAUGGUCAUCUUGGCGGCUGUUUUUC______AAAUCC___G_UU____________ACUG______UGGCAGU ..(((......(((((((....).)))))).(((((((...((((....)).))...))))))).................))).................................... (-24.35 = -24.80 + 0.45)


| Location | 17,873,112 – 17,873,217 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.72 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -30.63 |
| Energy contribution | -30.47 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17873112 105 - 22224390 ACUGCCA------CAGUUUCUUCGAUCGGAACC---GGAUUU------GGGAAACAGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAU .......------..((((((..(((((....)---.)))).------.)))))).(((((((...((.((....))))...))))))).((((((.(....)))))))........... ( -40.60) >DroPse_CAF1 144879 90 - 1 ACUGCCA------CAGU---------------C---AGAGUG------CAAAGAGAGCCGCCAAGAUGACUAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAU .((((((------((((---------------(---(....(------(..........)).....))))..)))))))).((((..(..((((((.(....)))))))...)...)))) ( -31.80) >DroWil_CAF1 164576 100 - 1 ACUGCCA------CUUUU-----------AAAA---GGAUUAAAGGGUAAAAAAAAGCCGCCAAGAUGACCAUGUGGCAGAGUGGUGGUAUGGGAGGCCAUGGCUCCCAAAAUAAUCCAU .......------.....-----------....---((((((..............(((((((...((.((....))))...))))))).((((((.(....)))))))...)))))).. ( -33.20) >DroYak_CAF1 132415 111 - 1 ACUGCCACAGACACAGUUUCUUCGAUCGGAAAC---GGAUUU------GGGAAACAGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAU .(((...))).....((((((..(((((....)---.)))).------.)))))).(((((((...((.((....))))...))))))).((((((.(....)))))))........... ( -43.20) >DroAna_CAF1 145779 96 - 1 ---------------------UCGAUCCGAGGCCAAGGAGGU---GACGACCAACAGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAU ---------------------..((.(((.((((..((.(..---..)..))..(((((((((...((.((....))))...))))))).))...)))).))).)).............. ( -37.20) >DroPer_CAF1 148743 90 - 1 ACUGCCA------CAGU---------------C---AGAGUG------CAAAGAGAGCCGCCAAGAUGACUAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAU .((((((------((((---------------(---(....(------(..........)).....))))..)))))))).((((..(..((((((.(....)))))))...)...)))) ( -31.80) >consensus ACUGCCA______CAGU____________AA_C___GGAGUU______GAAAAACAGCCGCCAAGAUGACCAUGUGGCAGAGUGGCGGCAUGGGAGGCCAUGGCUCCCAGAACAAUCCAU ....................................(((.................(((((((...((.((....))))...))))))).((((((.(....)))))))......))).. (-30.63 = -30.47 + -0.16)

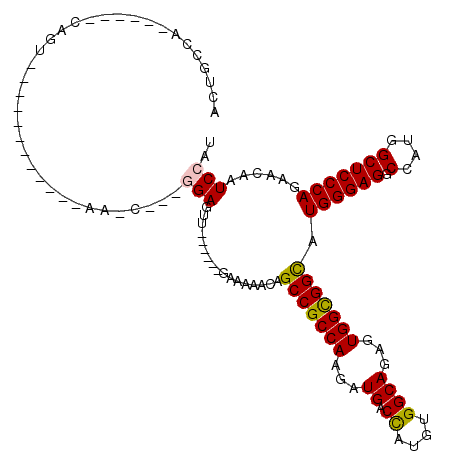

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:44 2006