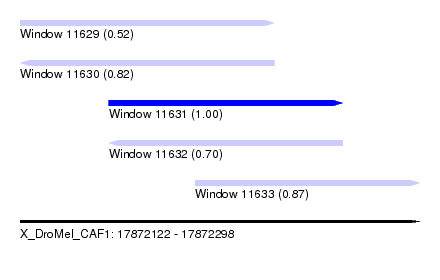
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,872,122 – 17,872,298 |
| Length | 176 |
| Max. P | 0.999766 |

| Location | 17,872,122 – 17,872,234 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 69.26 |
| Mean single sequence MFE | -39.78 |
| Consensus MFE | -7.61 |
| Energy contribution | -9.45 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.19 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17872122 112 + 22224390 AUUUAUAGCCAAUU-UAAAAGCCCAAAGAGCUGCACUUUCCGAGACCGUCGGCGCAUAGUGGUGGCCACUGGUGGCCACUGUUGGCUACUGGUAUCUACUGGUGGC-----CACUGGU .......((((...-....(((.......))).((((..((((.....)))).....))))(((((((....)))))))...(((((((..((....))..)))))-----)).)))) ( -39.80) >DroVir_CAF1 187014 106 + 1 AUUUAUAACCAAUUUUAAAAGCCCAAAGAGCCGCACCGACAGACGUCCGCUGC---CAGAAGCAGUGGGUGGUG-CCAUAG-----UGCAGUGGUGCAGUGCUG-CGCUGUAGCGG-- ........((...................(((((.(((((....))).(((((---.....))))))))))))(-(.((((-----((((((........))))-)))))).))))-- ( -36.50) >DroSec_CAF1 135623 92 + 1 AUUUAUAGCCAAUU-UAAAAGCCCAAAGAGCUGCACUUUCCGAGACCGUCGGCGCGUAGUGGCGGCCACUGGUGGCC--------------------ACUGGUGGC-----UACUGGU .......((((...-.....(((......(((((.(...((((.....)))).).))))))))((((((..((....--------------------))..)))))-----)..)))) ( -31.30) >DroSim_CAF1 106601 112 + 1 AUUUAUAGCCAAUU-UAAAAGCCCAAAGAGCUGCACUUUCCGAGACCGUCGGCGCAUAGUGGCGGCCACUGGUGGCCACUGUUGGCUACUGGUGGCCACUGGUGGC-----UACUGCU .....((((((...-..............((..(.....((((.....))))......)..))((((((..(((((((....)))))))..)))))).....))))-----))..... ( -46.90) >DroEre_CAF1 132629 109 + 1 AUUUAUAGCCAAUU-UAAAAGCCCAGAGGGCUGCACUUUCCGAAACUGGUGGUC---ACUAGUCGCUACUGCUGGCCACUGCUGGCCACUGCUGGCCACUGCUGGC-----CACUGGU ..............-....(((((...)))))............((..((((((---(.((((.((((..((((((((....))))))..)))))).)))).))))-----)))..)) ( -41.40) >DroYak_CAF1 131539 99 + 1 AUUUAUAGCCAAUU-UAAAAGCCCAAAGAGCUGCACUUUCCGAGACUGGCGGCC---ACU----------GAUGGCCACUGGUGGCCACUGGUGGCUACUGGUCUC-----CUCUGGU .......((((...-....(((.......))).........(((((..(..(((---(((----------..((((((....))))))..))))))..)..)))))-----...)))) ( -42.80) >consensus AUUUAUAGCCAAUU_UAAAAGCCCAAAGAGCUGCACUUUCCGAGACCGGCGGCG__UAGUGGCGGCCACUGGUGGCCACUG_UGGCUACUGGUGGCCACUGGUGGC_____CACUGGU .......((((..................(((((.....(((.......)))......))))).........((((((....))))))...........))))............... ( -7.61 = -9.45 + 1.84)


| Location | 17,872,122 – 17,872,234 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.26 |
| Mean single sequence MFE | -37.43 |
| Consensus MFE | -13.51 |
| Energy contribution | -13.57 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17872122 112 - 22224390 ACCAGUG-----GCCACCAGUAGAUACCAGUAGCCAACAGUGGCCACCAGUGGCCACCACUAUGCGCCGACGGUCUCGGAAAGUGCAGCUCUUUGGGCUUUUA-AAUUGGCUAUAAAU ....(((-----((((....((((..((((.(((.....(((((((....)))))))((((.....((((.....))))..))))..)))..))))...))))-...))))))).... ( -37.70) >DroVir_CAF1 187014 106 - 1 --CCGCUACAGCG-CAGCACUGCACCACUGCA-----CUAUGG-CACCACCCACUGCUUCUG---GCAGCGGACGUCUGUCGGUGCGGCUCUUUGGGCUUUUAAAAUUGGUUAUAAAU --..((((.((.(-(((..........)))).-----)).)))-)((((((((((((..(((---(((((....).))))))).)))).....))))..........))))....... ( -31.20) >DroSec_CAF1 135623 92 - 1 ACCAGUA-----GCCACCAGU--------------------GGCCACCAGUGGCCGCCACUACGCGCCGACGGUCUCGGAAAGUGCAGCUCUUUGGGCUUUUA-AAUUGGCUAUAAAU ....(((-----((((...((--------------------(((((....))))))).....(((.((((.....))))...))).((((.....))))....-...))))))).... ( -32.80) >DroSim_CAF1 106601 112 - 1 AGCAGUA-----GCCACCAGUGGCCACCAGUAGCCAACAGUGGCCACCAGUGGCCGCCACUAUGCGCCGACGGUCUCGGAAAGUGCAGCUCUUUGGGCUUUUA-AAUUGGCUAUAAAU ....(((-----((((...((((((((..((.((((....)))).))..))))))))((((.....((((.....))))..)))).((((.....))))....-...))))))).... ( -42.10) >DroEre_CAF1 132629 109 - 1 ACCAGUG-----GCCAGCAGUGGCCAGCAGUGGCCAGCAGUGGCCAGCAGUAGCGACUAGU---GACCACCAGUUUCGGAAAGUGCAGCCCUCUGGGCUUUUA-AAUUGGCUAUAAAU ....(((-----(((((((.(..((.((..((((((....))))))))....(.((((.((---....)).)))).)))..).)))(((((...)))))....-...))))))).... ( -38.70) >DroYak_CAF1 131539 99 - 1 ACCAGAG-----GAGACCAGUAGCCACCAGUGGCCACCAGUGGCCAUC----------AGU---GGCCGCCAGUCUCGGAAAGUGCAGCUCUUUGGGCUUUUA-AAUUGGCUAUAAAU .((((..-----(((((..(..(((((..(((((((....))))))).----------.))---)))..)..))))).((((((.(((....))).)))))).-..))))........ ( -42.10) >consensus ACCAGUG_____GCCACCAGUGGCCACCAGUAGCCA_CAGUGGCCACCAGUGGCCGCCACUA__CGCCGACGGUCUCGGAAAGUGCAGCUCUUUGGGCUUUUA_AAUUGGCUAUAAAU .......................................(((((((....................(((.......)))(((((.(((....))).)))))......))))))).... (-13.51 = -13.57 + 0.06)

| Location | 17,872,161 – 17,872,264 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -47.40 |
| Consensus MFE | -32.09 |
| Energy contribution | -35.78 |
| Covariance contribution | 3.69 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.68 |
| SVM decision value | 4.03 |
| SVM RNA-class probability | 0.999766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
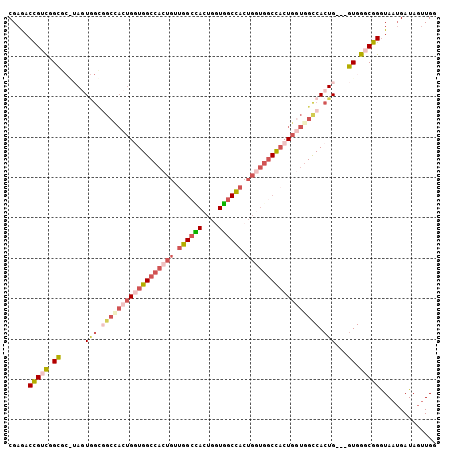
>X_DroMel_CAF1 17872161 103 + 22224390 CGAGACCGUCGGCGCAUAGUGGUGGCCACUGGUGGCCACUGUUGGCUACUGGUAUCUACUGGUGGCCACUGGUGGCCACUG---GUGGGCGGAUAAUGAUAGUUGG .....((((((((.(((....))))))((..(((((((((..(((((((..((....))..)))))))..)))))))))..---)).))))).............. ( -56.20) >DroSec_CAF1 135662 83 + 1 CGAGACCGUCGGCGCGUAGUGGCGGCCACUGGUGGCC--------------------ACUGGUGGCUACUGGUAGUCACUG---GUGGGCGGGUAAAGAUAGUUGG .....(((((.((....((((((.(((...))).)))--------------------)))((((((((....)))))))).---)).))))).............. ( -36.80) >DroSim_CAF1 106640 103 + 1 CGAGACCGUCGGCGCAUAGUGGCGGCCACUGGUGGCCACUGUUGGCUACUGGUGGCCACUGGUGGCUACUGCUGGUCACUG---GUGGGCGGGUAAUGAUAGUUGG .....((((((.(((...))).)((((((..(((((((....)))))))..))))))((..(((((((....)))))))..---)).))))).............. ( -52.60) >DroEre_CAF1 132668 100 + 1 CGAAACUGGUGGUC---ACUAGUCGCUACUGCUGGCCACUGCUGGCCACUGCUGGCCACUGCUGGCCACUGGUGGCCACUG---AUGGGCGGGUAAUGAUAGCUGG ....((((((....---)))))).((((.((((.(((.....((((((((..((((((....))))))..))))))))...---...))).))))....))))... ( -44.70) >DroYak_CAF1 131578 93 + 1 CGAGACUGGCGGCC---ACU----------GAUGGCCACUGGUGGCCACUGGUGGCUACUGGUCUCCUCUGGUGGCCACUGGCGGUGGGCGGGUAAUGAUAGUUGG .(((((..(..(((---(((----------..((((((....))))))..))))))..)..))))).((((.(.(((......))).).))))............. ( -46.70) >consensus CGAGACCGUCGGCGC_UAGUGGCGGCCACUGGUGGCCACUGUUGGCCACUGGUGGCCACUGGUGGCCACUGGUGGCCACUG___GUGGGCGGGUAAUGAUAGUUGG .....(((((.((......(((.((((((((((((((((((.((((((....)))))).)))))))))))))))))).)))...)).))))).............. (-32.09 = -35.78 + 3.69)


| Location | 17,872,161 – 17,872,264 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -11.28 |
| Energy contribution | -13.98 |
| Covariance contribution | 2.70 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
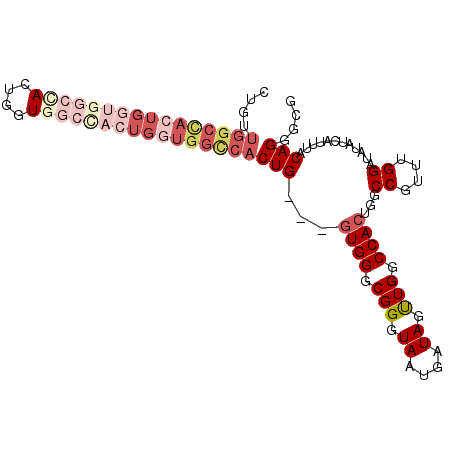
>X_DroMel_CAF1 17872161 103 - 22224390 CCAACUAUCAUUAUCCGCCCAC---CAGUGGCCACCAGUGGCCACCAGUAGAUACCAGUAGCCAACAGUGGCCACCAGUGGCCACCACUAUGCGCCGACGGUCUCG .........((..((.((.((.---..((((((((..((((((((..((.(.((....))..).)).))))))))..)))))))).....)).)).))..)).... ( -35.50) >DroSec_CAF1 135662 83 - 1 CCAACUAUCUUUACCCGCCCAC---CAGUGACUACCAGUAGCCACCAGU--------------------GGCCACCAGUGGCCGCCACUACGCGCCGACGGUCUCG ................(.((((---..(((.(((....))).)))..))--------------------)).)(((..((((((......)).))))..))).... ( -21.30) >DroSim_CAF1 106640 103 - 1 CCAACUAUCAUUACCCGCCCAC---CAGUGACCAGCAGUAGCCACCAGUGGCCACCAGUAGCCAACAGUGGCCACCAGUGGCCGCCACUAUGCGCCGACGGUCUCG ......................---..(.((((.......(((((..((((((((..((.....)).))))))))..)))))(((......))).....)))).). ( -33.90) >DroEre_CAF1 132668 100 - 1 CCAGCUAUCAUUACCCGCCCAU---CAGUGGCCACCAGUGGCCAGCAGUGGCCAGCAGUGGCCAGCAGUGGCCAGCAGUAGCGACUAGU---GACCACCAGUUUCG ...(((((..((..((((....---...((((((....))))))((..((((((....)))))))).))))..))..)))))((((.((---....)).))))... ( -33.90) >DroYak_CAF1 131578 93 - 1 CCAACUAUCAUUACCCGCCCACCGCCAGUGGCCACCAGAGGAGACCAGUAGCCACCAGUGGCCACCAGUGGCCAUC----------AGU---GGCCGCCAGUCUCG ................(.((((.....)))).).......(((((..(..(((((..(((((((....))))))).----------.))---)))..)..))))). ( -37.20) >consensus CCAACUAUCAUUACCCGCCCAC___CAGUGGCCACCAGUGGCCACCAGUAGCCACCAGUAGCCAACAGUGGCCACCAGUGGCCACCACUA_GCGCCGACGGUCUCG ...........................(.((((....((((((((.....((((....)))).....)))))))).(((((...)))))..........)))).). (-11.28 = -13.98 + 2.70)



| Location | 17,872,199 – 17,872,298 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -23.30 |
| Energy contribution | -27.30 |
| Covariance contribution | 4.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
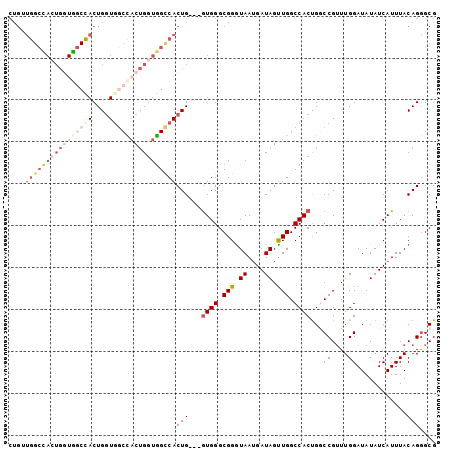
>X_DroMel_CAF1 17872199 99 + 22224390 CUGUUGGCUACUGGUAUCUACUGGUGGCCACUGGUGGCCACUG---GUGGGCGGAUAAUGAUAGUUGGCCACUGGCCGUUUGGAUAUAUCAUUUACAGGGCG ......(((.(((...(((((..(((((((....)))))))..---)))))..((((....(((.(((((...))))).)))....)))).....)))))). ( -39.40) >DroSec_CAF1 135699 80 + 1 -------------------ACUGGUGGCUACUGGUAGUCACUG---GUGGGCGGGUAAAGAUAGUUGGCCACUGGCCGUUUGGAUAUAUCAUUUACAGGGCG -------------------((..(((((((....)))))))..---))..((..(((((((((..(((((...)))))........)))).)))))...)). ( -27.10) >DroSim_CAF1 106678 99 + 1 CUGUUGGCUACUGGUGGCCACUGGUGGCUACUGCUGGUCACUG---GUGGGCGGGUAAUGAUAGUUGGCCACUGGCCGUUUGGAUAUAUCAUUUACAGGGCG ......(((.(((.((.((((..(((((((....)))))))..---)))).))...(((((((..(((((...)))))........)))))))..)))))). ( -37.40) >DroEre_CAF1 132703 99 + 1 CUGCUGGCCACUGCUGGCCACUGCUGGCCACUGGUGGCCACUG---AUGGGCGGGUAAUGAUAGCUGGCCACCGCCCGUUUGGAUAUAUCAUUUACAGAGCG ..((((((((((..((((((....))))))..))))))).(.(---((((((((.................))))))))).)................))). ( -44.03) >DroYak_CAF1 131603 102 + 1 CUGGUGGCCACUGGUGGCUACUGGUCUCCUCUGGUGGCCACUGGCGGUGGGCGGGUAAUGAUAGUUGGCCACUGACCGGUUGGAUAUAUCAUUUACAGGGCG (((.(.(((.(..((((((((..(......)..))))))))..).))).).)))(((((((((.(..(((.......)))..)...))))).))))...... ( -45.90) >consensus CUGUUGGCCACUGGUGGCCACUGGUGGCCACUGGUGGCCACUG___GUGGGCGGGUAAUGAUAGUUGGCCACUGGCCGUUUGGAUAUAUCAUUUACAGGGCG ....((((((((((((((((....))))))))))))))))(((...((((.(((.((....)).))).))))...((....))............))).... (-23.30 = -27.30 + 4.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:42 2006