
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,864,258 – 17,864,365 |
| Length | 107 |
| Max. P | 0.582972 |

| Location | 17,864,258 – 17,864,365 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.97 |
| Mean single sequence MFE | -23.34 |
| Consensus MFE | -11.65 |
| Energy contribution | -12.15 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
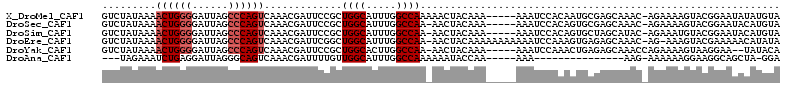
>X_DroMel_CAF1 17864258 107 - 22224390 GUCUAUAAAACUGGGGAUUAGCCCAGUCAAACGAUUCCGCUGGCAUUUGGCCAAAAACUACAAA-----AAAUCCACAAUGCGAGCAAAC-AGAAAAGUACGGAAUAUAUGUA .........((((((......))))))......((((((((.((((((((..............-----....))).))))).)))..((-......))..)))))....... ( -25.27) >DroSec_CAF1 121759 106 - 1 GUCUAUAAAACUGGGGAUUAGCCCAGUCAAACGAUUCCGCUGGCAUUUGGCCAA-AACUACAAA-----AAAUCCACAGUGCGAGCAAAC-AGAAAAGUACGGAAUACAUGUA .........((((((......))))))......((((((((.((((((((....-.........-----....))).))))).)))..((-......))..)))))....... ( -25.33) >DroSim_CAF1 98907 106 - 1 GUCUAUAAAACUGGGGAUUAGCCCAGUCAAACGAUUCCGCUGGCAUUUGGCCAA-AACUACAAA-----AAAUCCACAGUGCUAGCAUAC-AGAAAUGUACGGAAUACAUGUA .........((((((......))))))......(((((((((((((((((....-.........-----....))).))))))))).(((-(....)))).)))))....... ( -32.03) >DroEre_CAF1 125134 110 - 1 GUCUAUAAAACUGGGGAUUAGCCCAGUCAAACGAUUCGGCUGGCAUUUGGCCAA-AACUACAAAAAAAAAAAUCCAAAGUGAGAGCAAAC-AG-AAAGUACGAAAAACAUAUA ..........(((.(((((((((.((((....)))).))))(((.....)))..-...............)))))...((....))...)-))-................... ( -23.00) >DroYak_CAF1 123653 105 - 1 GUCUAUAAAACUGGGGAUUAGCCCAGUCAAACGAUUCCGCUGGCACUUGGCCAA-AACUACAAA-----AAAUCCAAACUGAGAGCAAACCAGAAAAGUAAGGAA--UAUACA .........((((((......))))))......(((((..((((.....)))).-.........-----.........(((.(......))))........))))--)..... ( -20.50) >DroAna_CAF1 135299 88 - 1 ---UAGAAAUCUGAGGAUUAGGGCAGUCAAACGAUUUUGUUGGCAUUUGGCCAAAAAAUACCAA-----AAA---------------AAG-AAAAAAGGAAGGCAGCUA-GGA ---....((((....))))..(((.(((.....(((((.(((((.....)))))))))).((..-----...---------------...-......))..))).))).-... ( -13.94) >consensus GUCUAUAAAACUGGGGAUUAGCCCAGUCAAACGAUUCCGCUGGCAUUUGGCCAA_AACUACAAA_____AAAUCCACAGUGCGAGCAAAC_AGAAAAGUACGGAAUACAUGUA .........((((((......)))))).............((((.....))))............................................................ (-11.65 = -12.15 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:35 2006