| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,861,890 – 17,861,994 |
| Length | 104 |
| Max. P | 0.615026 |

| Location | 17,861,890 – 17,861,994 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 65.13 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -9.12 |
| Energy contribution | -8.98 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
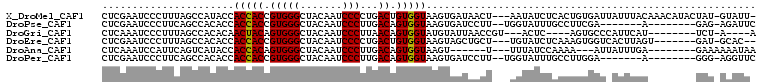
>X_DroMel_CAF1 17861890 104 + 22224390 CUCGAAUCCCUUUAGCCAUACCACCACCGUGGGCUACAAUCCCCUGACUGUGGUAAGUGAUAACU---AAUAUCUCACUGUGAUUAUUUACAAACAUACUAU-GUAUU- ..................((((((...((.(((.......))).))...))))))(((((.....---......)))))((((....))))...........-.....- ( -15.90) >DroPse_CAF1 134161 91 + 1 CUCGAAUCCCUUCAGCCACACCACCACCGUGGGCUACAAUCCCUUGACAGUGGUAAGUGAUCCUU--UGGUAUUUGCCUUCGA-------A--------GAG-AGAUUC ...((((((((((((((.(((.......)))))))................(((((((.(((...--.))))))))))...))-------)--------).)-.))))) ( -23.50) >DroGri_CAF1 148759 89 + 1 CUCAAAUCCCUUUAGCCACACAACUACAGUGGGCUACAAUCCCUUAACAGUGGUAUGUAUUAACCGU---ACUC----AGUGCCCAUUCAU--------UCU-A----A ............(((((.(((.......))))))))...........((.(((((((.......)))---)).)----).)).........--------...-.----. ( -16.10) >DroEre_CAF1 123028 96 + 1 CUCGAAUCCCUUUAGCCACACCACCACCGUGGGCUACAAUCCCCUGACUGUGGUAAGUAGCUGCU---UGUAUCUCAAAGUGGUCACUUAGU-------GAU-GCAC-- ............((((.((.((((....))))((((((.((....)).))))))..)).))))..---((((((.(.((((....)))).).-------)))-))).-- ( -25.50) >DroAna_CAF1 133081 89 + 1 CUCAAAUCCAUUCAGUCAUACCACCACAGUGGGCUACAAUCCCUUGACAGUGGUAAGU------U---UUUAUCCAAAA---AUUAUUUGA--------GAAAAAAUAA (((((((...........((((((..(((.(((.......))))))...))))))(((------(---(((....))))---)))))))))--------)......... ( -18.20) >DroPer_CAF1 136048 91 + 1 CUCGAAUCCCUUCAGCCACACCACCACCGUGGGCUACAAUCCCUUGACAGUGGUAAGUGAUCCUU--UGGUAUUUGCCUUGGA-------A--------GGG-AGGUUC ......(((((((((((.(((.......)))))))............(((.(((((((.(((...--.)))))))))))))))-------)--------)))-)..... ( -28.30) >consensus CUCGAAUCCCUUCAGCCACACCACCACCGUGGGCUACAAUCCCUUGACAGUGGUAAGUGAUAACU___GGUAUCUCACAGUGAU_AUUUAA________GAU_AGAUU_ ......................(((((.(((((.......)))...)).)))))....................................................... ( -9.12 = -8.98 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:34 2006