| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,860,420 – 17,860,540 |
| Length | 120 |
| Max. P | 0.532004 |

| Location | 17,860,420 – 17,860,540 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.94 |
| Mean single sequence MFE | -41.49 |
| Consensus MFE | -27.79 |
| Energy contribution | -28.52 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
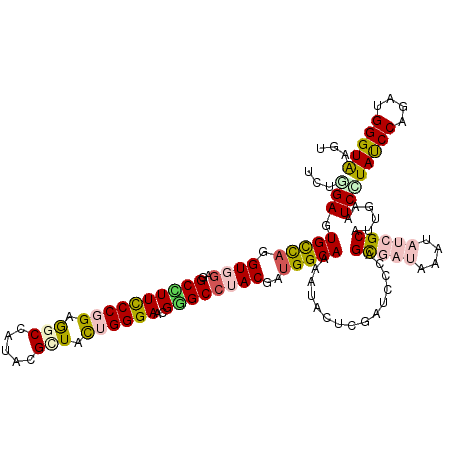
>X_DroMel_CAF1 17860420 120 + 22224390 ACUCGAGUGUUUGGUGGAGGCAUUCCCCGAAGCCAUUAGGUAUUGGGAACGUGCCUACGACGGCAAGAUCCUCGAUCCCAGCGACAAGUACGGCAUUGAAUCGUACCCAGAGGGGUGAGU (((((..(.((((((((((((((..(((((.(((....))).)))))...)))))).(((.((......)))))...))).......((((((.......))))))))))).)..))))) ( -40.60) >DroVir_CAF1 172075 120 + 1 AUUGGAAUGCCAGGUGGAGGCCUUCCCGGAGGCCAUACGCUACUGGGAGCGCGCCUACGAUGGCAAGAUUCUCGAUCCAAGUGAUAAGUAUCGCAUUGAAUCAUAUCCAGACGGGUGAGU .(((((.(((((.((((..((.(((((((.(((.....))).))))))).))..))))..)))))........(((.((((((((....))))).))).)))...))))).......... ( -42.50) >DroGri_CAF1 147204 120 + 1 UUUGGAGUGCCAGGUGGAAGCUUUCCCGGAGGCCAUACGCUACUGGGAGCGGGCCUACGAUGGCAAAAUUCUCGAGCCGAGUGAUAAAUAUCGCAUCGAGUCCUAUCCAGAUGGGUAUGU ...(((.(((((.((((..((((((((((.(((.....))).)))))))..)))))))..))))).....(((((.....(((((....))))).))))))))(((((....)))))... ( -42.20) >DroWil_CAF1 151219 120 + 1 UCUGGAAUGUCAGGUUGAGGCCUUCCCCGAGGCGAUACGCUAUUGGGAACGGGCAUACGAUGGCAAAAUAUUAGAUCCCAGCGAUAAAUAUCGCAUCGAAUCGUAUCCCGAUGGGUGGGU (((((....))))).....((((..(((((((((...)))).)))))..((((.(((((((((..............)).(((((....))))).....)))))))))))..)))).... ( -35.94) >DroMoj_CAF1 187618 120 + 1 CCUGGAGUGCCAGGUCGAAGCCUUUCCGGAGGCUAUACGGUACUGGGAGCGGGCCUACGAUGGCAAAAUAUUGGAUCCAAGUGAUAAGUAUCGCAUUGAGUCCUAUCCAGAUGGGUAAGU .(((((.(((((..(((.((((((....))))))....(((.(((....))))))..)))))))).......(((..((((((((....))))).)))..)))..))))).......... ( -41.30) >DroAna_CAF1 131582 120 + 1 UUUGGAGUGCCAAGUGGAGGCCUUCCCGGAGGCCAUUCGUUACUGGGAAAGGGCCUACGAUGGCAAGAUCCUAGAUCCCAGCGACAAAUACGGCAUAGAAUCUUAUCCAGAUGGGUAGGU ......(((((....(((((((((....)))))).)))((..((((((.((((((......))).....)))...))))))..))......))))).....(((((((....))))))). ( -46.40) >consensus UCUGGAGUGCCAGGUGGAGGCCUUCCCGGAGGCCAUACGCUACUGGGAACGGGCCUACGAUGGCAAAAUACUCGAUCCCAGCGAUAAAUAUCGCAUUGAAUCCUAUCCAGAUGGGUAAGU ...(((.(((((.((((..((((((((((.(((.....))).))))))..))))))))..)))))...............(((((....)))))......)))(((((....)))))... (-27.79 = -28.52 + 0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:33 2006