
| Sequence ID | X_DroMel_CAF1 |
|---|---|
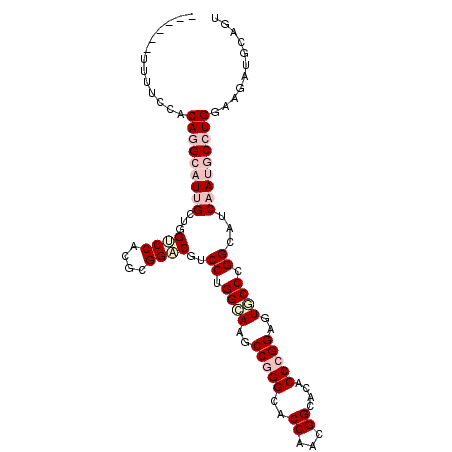
| Location | 17,859,751 – 17,859,849 |
| Length | 98 |
| Max. P | 0.897572 |

| Location | 17,859,751 – 17,859,849 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.70 |
| Mean single sequence MFE | -40.45 |
| Consensus MFE | -26.83 |
| Energy contribution | -28.83 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
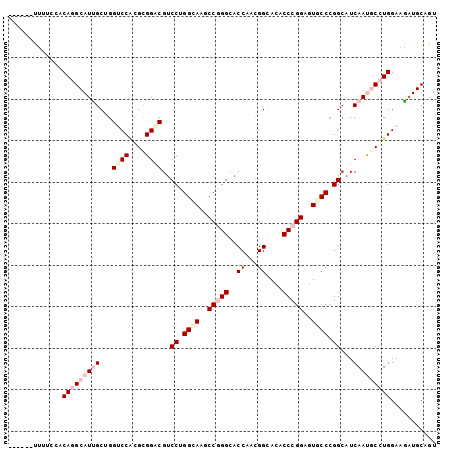
>X_DroMel_CAF1 17859751 98 + 22224390 -----CUUCUGGACAGGCAUUGCGGGUCCACGUGGUCGACCUGGCAAGCCGGGAACCAAUGGCAUACCCGGAGUGCCCGGCAUCAAUGCCUGGAAGCUGCAAU -----(((((.....((((((.((((((((..((((...(((((....))))).)))).)))...))))).)))))).((((....)))).)))))....... ( -42.40) >DroVir_CAF1 169761 103 + 1 CCCUGCUCAAGCACAGGCAUCGCCGGGCCACGCGGACGUCCUGGCAAGCCGGGCACCAAUGGCACACCCGGCGUGCCGGGCAUCAACACCUGGAAGAUGCAAU ...(((....)))...(((((.(((((((....))..(.(((((((.((((((..((...))....)))))).)))))))).......)))))..)))))... ( -44.00) >DroPse_CAF1 131998 97 + 1 ------CUUUCCACAGGCAUUGCUGGUCCGCGCGGACGUCCUGGAAAGCCGGGCACCAACGGUACACCCGGAGUGCCCGGCAUCAACGCCUGGAAGAUGCAGU ------.((((((..(((.(((...((((....))))..........(((((((((...(((.....)))..)))))))))..))).)))))))))....... ( -42.10) >DroGri_CAF1 146462 98 + 1 -----CUUAAUUGCAGGCAUUGCUGGUCCACGCGGACGUCCGGGCAAGCCAGGCACCAAUGGCACACCGGGUGUGCCCGGCAUCAAUACGUGGAAAAUGCAGU -----....(((((((((...)))..((((((..((.(.((((((..((((........))))((((...))))))))))).))....))))))...)))))) ( -37.90) >DroAna_CAF1 130627 97 + 1 ------UUUCCUACAGGCAUCGCAGGACCUCGCGGUCGUCCUGGCAAGCCCGGCACCAACGGCACUCCCGGAGUCCCCGGAAUCAAUGCCUGGAAGAUGCAGU ------..........(((((....((((....)))).(((.((((.(((.(.......))))....((((.....))))......)))).))).)))))... ( -34.00) >DroPer_CAF1 133856 97 + 1 ------CUUUCCACAGGCAUUGCUGGUCCGCGCGGACGUCCUGGAAAGCCGGGCACCAACGGCACGCCCGGAGUUCCCGGCAUCAAUGCAUGGAAAAUGCAGU ------.((((((...((((((.((((((....))))..((.((((..((((((...........))))))..)))).)))).)))))).))))))....... ( -42.30) >consensus ______UUUUCCACAGGCAUUGCUGGUCCACGCGGACGUCCUGGCAAGCCGGGCACCAACGGCACACCCGGAGUGCCCGGCAUCAAUGCCUGGAAGAUGCAGU .............(((((((((...((((....))))..((.((((..(((((..((...))....)))))..)))).))...)))))))))........... (-26.83 = -28.83 + 2.00)

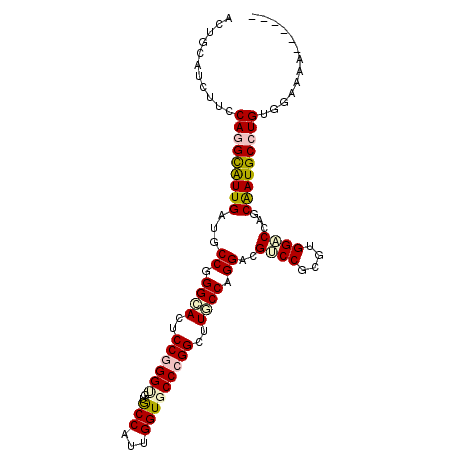
| Location | 17,859,751 – 17,859,849 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.70 |
| Mean single sequence MFE | -44.42 |
| Consensus MFE | -32.99 |
| Energy contribution | -33.43 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17859751 98 - 22224390 AUUGCAGCUUCCAGGCAUUGAUGCCGGGCACUCCGGGUAUGCCAUUGGUUCCCGGCUUGCCAGGUCGACCACGUGGACCCGCAAUGCCUGUCCAGAAG----- .......(((((((((((((......((((..(((((...(((...))).)))))..)))).((((.((...)).))))..)))))))))....))))----- ( -41.50) >DroVir_CAF1 169761 103 - 1 AUUGCAUCUUCCAGGUGUUGAUGCCCGGCACGCCGGGUGUGCCAUUGGUGCCCGGCUUGCCAGGACGUCCGCGUGGCCCGGCGAUGCCUGUGCUUGAGCAGGG (((((.....(((.(((..((((((.((((.(((((((..(((...)))))))))).)))).)).))))))).)))....))))).(((((......))))). ( -49.90) >DroPse_CAF1 131998 97 - 1 ACUGCAUCUUCCAGGCGUUGAUGCCGGGCACUCCGGGUGUACCGUUGGUGCCCGGCUUUCCAGGACGUCCGCGCGGACCAGCAAUGCCUGUGGAAAG------ ........((((((((((((..((((((((((.(((.....)))..))))))))))..........((((....))))...))))))))).)))...------ ( -47.70) >DroGri_CAF1 146462 98 - 1 ACUGCAUUUUCCACGUAUUGAUGCCGGGCACACCCGGUGUGCCAUUGGUGCCUGGCUUGCCCGGACGUCCGCGUGGACCAGCAAUGCCUGCAAUUAAG----- ..((((...(((((((...(((((((((((...((((.(..((...))..)))))..))))))).)))).))))))).((....))..))))......----- ( -43.50) >DroAna_CAF1 130627 97 - 1 ACUGCAUCUUCCAGGCAUUGAUUCCGGGGACUCCGGGAGUGCCGUUGGUGCCGGGCUUGCCAGGACGACCGCGAGGUCCUGCGAUGCCUGUAGGAAA------ ......((((.(((((((((..(((((.((..((((.(.(......).).))))..)).)).))).((((....))))...))))))))).))))..------ ( -41.90) >DroPer_CAF1 133856 97 - 1 ACUGCAUUUUCCAUGCAUUGAUGCCGGGAACUCCGGGCGUGCCGUUGGUGCCCGGCUUUCCAGGACGUCCGCGCGGACCAGCAAUGCCUGUGGAAAG------ .......(((((((((((((.((((.((((..((((((..(((...)))))))))..)))).))..((((....)))))).))))))..))))))).------ ( -42.00) >consensus ACUGCAUCUUCCAGGCAUUGAUGCCGGGCACUCCGGGUGUGCCAUUGGUGCCCGGCUUGCCAGGACGUCCGCGUGGACCAGCAAUGCCUGUGGAAAA______ ...........(((((((((...((.((((..((((((..(((...)))))))))..)))).))..((((....))))...)))))))))............. (-32.99 = -33.43 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:31 2006