
| Sequence ID | X_DroMel_CAF1 |
|---|---|
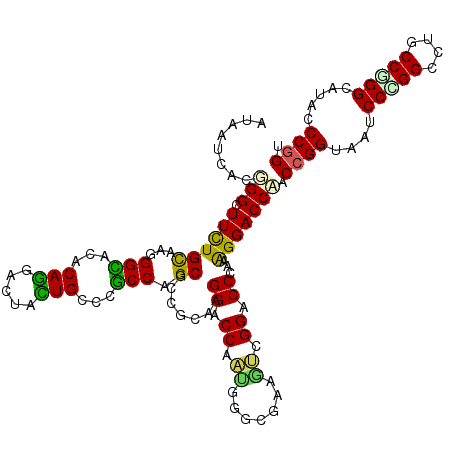
| Location | 17,859,269 – 17,859,389 |
| Length | 120 |
| Max. P | 0.963862 |

| Location | 17,859,269 – 17,859,389 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.00 |
| Mean single sequence MFE | -51.20 |
| Consensus MFE | -27.48 |
| Energy contribution | -26.40 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17859269 120 + 22224390 AUAAUCACGGGCUUCUGCAAGGGUACCCAGGACUAUUGUCCGCCGGCACCGCAAGGACCAACCGGAGAAGUGGGACCAAAGGGACCACCGGGAAACCCGGGUCUACCUGGCAUCCCCGGU .......((((....(((.(((((.((((((((....)))).((((..((....)).....)))).....)))))))....(((((.(.((....)).))))))..)).)))..)))).. ( -49.50) >DroVir_CAF1 169278 120 + 1 AUAAUUACCGGCUUUUGCAAGGGCACACAGGACUAUUGCCCGCCAGCGCCGGCCGGGCCAAUGGGCCCGAUUGGACCCAAAGGACCAACCGGUAAUCCCGGCCUGCCGGGCAUACCCGGU .......(((((.((.((..(((((...........))))))).)).))))).((((((....))))))..(((.((....)).)))(((((..(((((((....))))).))..))))) ( -53.00) >DroGri_CAF1 145979 120 + 1 AUAAUCACUGGCUUUUGCAAGGGCACACAGGAAUACUGCCCGCCAGCACCGUCAGGACCAACGGGUGCCGUCGGCCCAAAAGGACCACCCGGUAAUCCUGGCUUACCGGGCAUACCCGGC .......(((((........(((((...........))))))))))....(((((((....((((((..(((..........)))))))))....)))))))...(((((....))))). ( -45.20) >DroWil_CAF1 150095 120 + 1 AUUAUCACGGGAUUCUGUAAGGGUACUCAAGACUAUUGUCCACCUGCGCCGCAGGGACCACAAGGUGAUAUCGGACCGAAGGGACCCACCGGUAACCCCGGAUUGCCUGGAAUUCCCGGA .......(((((((((.....(((......(((....)))..(((((...))))).)))...(((..((...((.((....)).))..((((.....))))))..))))))).))))).. ( -42.20) >DroMoj_CAF1 186460 120 + 1 AUUAUCACGGGCUUUUGCAAGGGCACACAGGAAUACUGCCCGCCAGCCCCAGCUGGCCCAAUGGGCCCAGUAGGACCCAAAGGACCCACCGGCAAUCCCGGCCUGCCGGGCAUACCCGGU ........((((((((....(((((...........)))))((((((....)))))).....(((.((....)).)))))))).)))(((((..(((((((....))))).))..))))) ( -51.00) >DroAna_CAF1 130146 120 + 1 AUCAUCACGGGCUUCUGCAAGGGCACCCAGGACUACUGUCCGCCGGCGCCCCAGGGUCCGGUGGGCGAGGCGGGUCCCAAAGGACCCACCGGAAAUCCGGGUCUGCCGGGCAUACCCGGU .......((((..((((...((((.((..((((....))))...)).))))))))((((((..(((.....((((((....)))))).((((....)))))))..))))))...)))).. ( -66.30) >consensus AUAAUCACGGGCUUCUGCAAGGGCACACAGGACUACUGCCCGCCAGCGCCGCAAGGACCAAUGGGCGAAGUCGGACCCAAAGGACCAACCGGUAAUCCCGGCCUGCCGGGCAUACCCGGU ........(((.((((((...(((...(((.....)))...))).)).......((.((.((.......)).)).))...))))))).((((....(((((....))))).....)))). (-27.48 = -26.40 + -1.08)


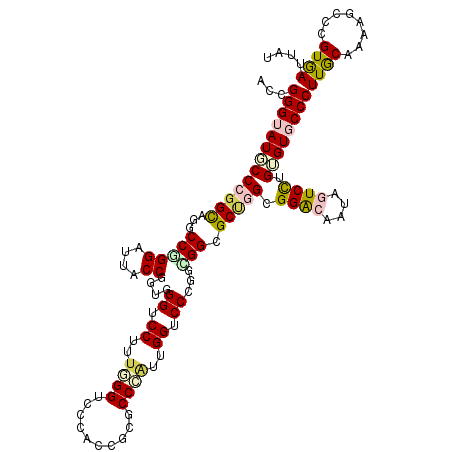
| Location | 17,859,269 – 17,859,389 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.00 |
| Mean single sequence MFE | -56.55 |
| Consensus MFE | -29.23 |
| Energy contribution | -29.78 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.941090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17859269 120 - 22224390 ACCGGGGAUGCCAGGUAGACCCGGGUUUCCCGGUGGUCCCUUUGGUCCCACUUCUCCGGUUGGUCCUUGCGGUGCCGGCGGACAAUAGUCCUGGGUACCCUUGCAGAAGCCCGUGAUUAU ((((((((.((((((..((((((((...))))..))))..))))))......)))))))).(((..((((((((((.(.((((....))))).))))))...))))..)))......... ( -49.90) >DroVir_CAF1 169278 120 - 1 ACCGGGUAUGCCCGGCAGGCCGGGAUUACCGGUUGGUCCUUUGGGUCCAAUCGGGCCCAUUGGCCCGGCCGGCGCUGGCGGGCAAUAGUCCUGUGUGCCCUUGCAAAAGCCGGUAAUUAU (((((...((((((.((((((((.....))((((((.((..(((((((....)))))))..)).))))))))).))).))))))......((...(((....)))..)))))))...... ( -65.10) >DroGri_CAF1 145979 120 - 1 GCCGGGUAUGCCCGGUAAGCCAGGAUUACCGGGUGGUCCUUUUGGGCCGACGGCACCCGUUGGUCCUGACGGUGCUGGCGGGCAGUAUUCCUGUGUGCCCUUGCAAAAGCCAGUGAUUAU ((((((..((((((((((.......))))))))))..))....((((((((((...))))))))))...))))(((((((((((.(((....))))))))((....))))))))...... ( -56.20) >DroWil_CAF1 150095 120 - 1 UCCGGGAAUUCCAGGCAAUCCGGGGUUACCGGUGGGUCCCUUCGGUCCGAUAUCACCUUGUGGUCCCUGCGGCGCAGGUGGACAAUAGUCUUGAGUACCCUUACAGAAUCCCGUGAUAAU (((((((..((.((((....(((((..(((....)))..)))))(((((.....(((....))).(((((...))))))))))....)))).))(((....)))....))))).)).... ( -38.40) >DroMoj_CAF1 186460 120 - 1 ACCGGGUAUGCCCGGCAGGCCGGGAUUGCCGGUGGGUCCUUUGGGUCCUACUGGGCCCAUUGGGCCAGCUGGGGCUGGCGGGCAGUAUUCCUGUGUGCCCUUGCAAAAGCCCGUGAUAAU .((((((...((((((..(((((.....))))).(((((..(((((((....)))))))..))))).))))))((.((((.((((.....)))).))))...))....))))).)..... ( -67.00) >DroAna_CAF1 130146 120 - 1 ACCGGGUAUGCCCGGCAGACCCGGAUUUCCGGUGGGUCCUUUGGGACCCGCCUCGCCCACCGGACCCUGGGGCGCCGGCGGACAGUAGUCCUGGGUGCCCUUGCAGAAGCCCGUGAUGAU .((((((.(((...))).))))))......(((((((((....)))))))))(((..(((.((...((((((((((.(.((((....))))).)))))))...)))...)).))).))). ( -62.70) >consensus ACCGGGUAUGCCCGGCAGGCCGGGAUUACCGGUGGGUCCUUUGGGUCCCACCGCGCCCAUUGGUCCCGGCGGCGCUGGCGGACAAUAGUCCUGUGUGCCCUUGCAAAAGCCCGUGAUUAU ...(((((((((((((...(((((....))....((.((..((((..........))))..)).))...))).))))).((((....)))).))))))))((((........)))).... (-29.23 = -29.78 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:30 2006