
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,856,196 – 17,856,471 |
| Length | 275 |
| Max. P | 0.993442 |

| Location | 17,856,196 – 17,856,286 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 87.94 |
| Mean single sequence MFE | -24.56 |
| Consensus MFE | -18.51 |
| Energy contribution | -19.14 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17856196 90 - 22224390 AGUACAAUGGCCAGAAAAGUGGGUGGAAUGAAGCCAUUCGCCGGCAAAGGCCAAUGUCAGAAAGUUGGCAGCAUCAG---GUGCACUGAAAGA .((((..(((((......((.((((.((((....)))))))).))...))))).((((((....)))))).......---))))......... ( -26.90) >DroSec_CAF1 113757 78 - 1 AAUACAAUGGCCAGAAAAGUGGGUGGAAUGAAGCCAUUUGCCGGCAAAGGCCAAUGUCAGAAAGUUGG---------------CACUGAAAGA .......(((((......((.((..((.((....))))..)).))...))))).((((((....))))---------------))........ ( -19.90) >DroSim_CAF1 90871 90 - 1 AAUACAAUGGCCAGAAAAGUGGGUGGAAUGAAGCCAUUUGCCGGCAAAGGCCAAUGUCAGAAAGUUGGCAGCAUCAG---GUGCACUGAAAGA .......(((((......((.((..((.((....))))..)).))...))))).((((((....))))))...((((---.....)))).... ( -24.20) >DroEre_CAF1 117457 93 - 1 AGUACAAUGGCCAGAAGAGUGGGCGGAAUGGAGCCAUUUGCCGUCAAAGGCCAAUGUCAGAAAGUUGGCAGCAUCUCUUGGUGCACUGAAAGA ....((.((((((..(((((.(((((.(((....)))...)))))....((((((........)))))).)).)))..)))).)).))..... ( -27.60) >DroYak_CAF1 115485 89 - 1 AGUAUAAUGGCCAGAAAAGUGGGUGGAAUGGAGCCAUUUGCCGUCACAGGCCAA-GUCAGAAAGUUGGCAGCAUCAU---GUGCACUGAAAGA .(((((.(((((.((...(..(((((.......)))))..)..))...))))).-(((((....))))).......)---))))......... ( -24.20) >consensus AGUACAAUGGCCAGAAAAGUGGGUGGAAUGAAGCCAUUUGCCGGCAAAGGCCAAUGUCAGAAAGUUGGCAGCAUCAG___GUGCACUGAAAGA .......(((((......((.((((.((((....)))))))).))...)))))..(((((....))))).((((......))))......... (-18.51 = -19.14 + 0.63)

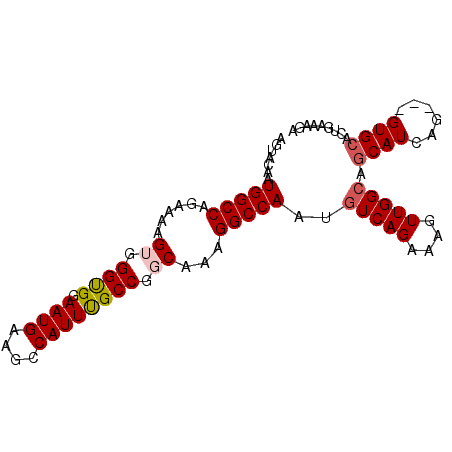
| Location | 17,856,226 – 17,856,325 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 93.54 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -26.80 |
| Energy contribution | -27.04 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17856226 99 + 22224390 CUGACAUUGGCCUUUGCCGGCGAAUGGCUUCAUUCCACCCACUUUUCUGGCCAUUGUACUUGGCCAACAAAUGCAGCGCACAGCUGGCAACAAAGUGAA ....((((.(((......))).)))).............((((((..((((((.......)))))).....((((((.....))).)))..)))))).. ( -30.80) >DroSec_CAF1 113775 99 + 1 CUGACAUUGGCCUUUGCCGGCAAAUGGCUUCAUUCCACCCACUUUUCUGGCCAUUGUAUUUGGCCAACUAAUGCAGCACACAGCUGGCAACAAAGUGAA ..((....((((((((....)))).))))....))....((((((..((((((.......)))))).....((((((.....))).)))..)))))).. ( -28.50) >DroSim_CAF1 90901 99 + 1 CUGACAUUGGCCUUUGCCGGCAAAUGGCUUCAUUCCACCCACUUUUCUGGCCAUUGUAUUUGGCCAACUAAUGCAGCACACAGCUGGCAACAAAGUGAA ..((....((((((((....)))).))))....))....((((((..((((((.......)))))).....((((((.....))).)))..)))))).. ( -28.50) >DroEre_CAF1 117490 99 + 1 CUGACAUUGGCCUUUGACGGCAAAUGGCUCCAUUCCGCCCACUCUUCUGGCCAUUGUACUUGGCCAACUAAUGCAGCGCACAGCUGGCAACAAGGUGAA (((.((((((........(((.((((....))))..)))........((((((.......)))))).)))))))))..(((...((....))..))).. ( -29.30) >DroYak_CAF1 115515 98 + 1 CUGAC-UUGGCCUGUGACGGCAAAUGGCUCCAUUCCACCCACUUUUCUGGCCAUUAUACUUGGCCAACUAAGGCAGCGCGCAGCUGGCAACAAAGUGAA .....-...(((......))).((((....)))).....((((((..((((((.......)))))).....(.((((.....)))).)...)))))).. ( -28.60) >consensus CUGACAUUGGCCUUUGCCGGCAAAUGGCUUCAUUCCACCCACUUUUCUGGCCAUUGUACUUGGCCAACUAAUGCAGCGCACAGCUGGCAACAAAGUGAA ....((((.(((......))).)))).............((((((..((((((.......)))))).......((((.....)))).....)))))).. (-26.80 = -27.04 + 0.24)


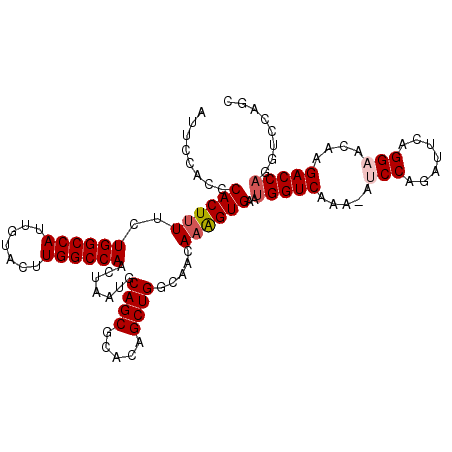
| Location | 17,856,257 – 17,856,364 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 89.42 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.84 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17856257 107 + 22224390 AUUCCACCCACUUUUCUGGCCAUUGUACUUGGCCAACAAAUGCAGCGCACAGCUGGCAACAAAGUGAAUGGUCAAA-AUCCAGAUUUAGGAACAAGACCAGGUCCAGC .....(((((((((..((((((.......)))))).....((((((.....))).)))..))))))..(((((...-.(((.......)))....))))))))..... ( -31.90) >DroSec_CAF1 113806 108 + 1 AUUCCACCCACUUUUCUGGCCAUUGUAUUUGGCCAACUAAUGCAGCACACAGCUGGCAACAAAGUGAAUGGUCAAAAACCCAGAUUCAGGAACAGGACCAGGUCCAGC .....(((((((((..((((((.......)))))).....((((((.....))).)))..))))))..(((((.....((........)).....))))))))..... ( -30.20) >DroSim_CAF1 90932 108 + 1 AUUCCACCCACUUUUCUGGCCAUUGUAUUUGGCCAACUAAUGCAGCACACAGCUGGCAACAAAGUGAAUGGUCAAAAACCCAGAUUCAGGAACAGGACCAGGUCCAGC .....(((((((((..((((((.......)))))).....((((((.....))).)))..))))))..(((((.....((........)).....))))))))..... ( -30.20) >DroEre_CAF1 117521 107 + 1 AUUCCGCCCACUCUUCUGGCCAUUGUACUUGGCCAACUAAUGCAGCGCACAGCUGGCAACAAGGUGAAUGGUCAAA-AUCCGGAUUCGGGAAUUAGACCAGGUCCAGC .....((((((.(((.((((((.......)))))).....((((((.....))).)))..))))))..(((((.((-.(((.......))).)).))))))))..... ( -32.00) >DroYak_CAF1 115545 98 + 1 AUUCCACCCACUUUUCUGGCCAUUAUACUUGGCCAACUAAGGCAGCGCGCAGCUGGCAACAAAGUGAAUGGUCAAA-AUCCAGAUUCAGGAACAAGACC--------- ........((((((..((((((.......)))))).....(.((((.....)))).)...))))))...((((...-.(((.......)))....))))--------- ( -29.10) >consensus AUUCCACCCACUUUUCUGGCCAUUGUACUUGGCCAACUAAUGCAGCGCACAGCUGGCAACAAAGUGAAUGGUCAAA_AUCCAGAUUCAGGAACAAGACCAGGUCCAGC ........((((((..((((((.......)))))).......((((.....)))).....))))))..(((((.....(((.......)))....)))))........ (-26.20 = -26.84 + 0.64)

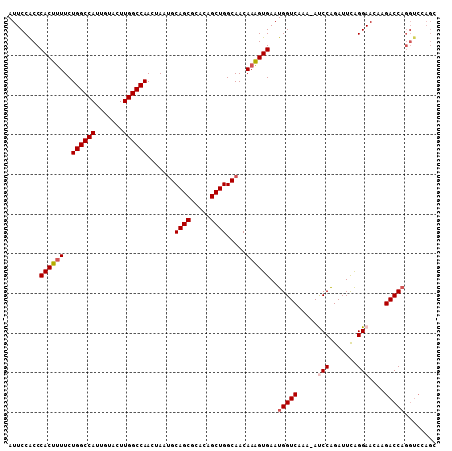
| Location | 17,856,257 – 17,856,364 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 89.42 |
| Mean single sequence MFE | -34.78 |
| Consensus MFE | -29.76 |
| Energy contribution | -29.96 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17856257 107 - 22224390 GCUGGACCUGGUCUUGUUCCUAAAUCUGGAU-UUUGACCAUUCACUUUGUUGCCAGCUGUGCGCUGCAUUUGUUGGCCAAGUACAAUGGCCAGAAAAGUGGGUGGAAU ......((.((((....(((.......))).-...))))(((((((((..(((.(((.....))))))....(((((((.......))))))).)))))))))))... ( -34.40) >DroSec_CAF1 113806 108 - 1 GCUGGACCUGGUCCUGUUCCUGAAUCUGGGUUUUUGACCAUUCACUUUGUUGCCAGCUGUGUGCUGCAUUAGUUGGCCAAAUACAAUGGCCAGAAAAGUGGGUGGAAU ...(((((..(..............)..)))))....(((((((((((..(((.(((.....))))))....(((((((.......))))))).)))))))))))... ( -36.14) >DroSim_CAF1 90932 108 - 1 GCUGGACCUGGUCCUGUUCCUGAAUCUGGGUUUUUGACCAUUCACUUUGUUGCCAGCUGUGUGCUGCAUUAGUUGGCCAAAUACAAUGGCCAGAAAAGUGGGUGGAAU ...(((((..(..............)..)))))....(((((((((((..(((.(((.....))))))....(((((((.......))))))).)))))))))))... ( -36.14) >DroEre_CAF1 117521 107 - 1 GCUGGACCUGGUCUAAUUCCCGAAUCCGGAU-UUUGACCAUUCACCUUGUUGCCAGCUGUGCGCUGCAUUAGUUGGCCAAGUACAAUGGCCAGAAGAGUGGGCGGAAU .(((((..(((........)))..)))))((-((((.((((((..((...(((.(((.....))))))..))(((((((.......)))))))..)))))).)))))) ( -35.50) >DroYak_CAF1 115545 98 - 1 ---------GGUCUUGUUCCUGAAUCUGGAU-UUUGACCAUUCACUUUGUUGCCAGCUGCGCGCUGCCUUAGUUGGCCAAGUAUAAUGGCCAGAAAAGUGGGUGGAAU ---------((((....(((.......))).-...))))(((((((((...(.((((.....)))).)....(((((((.......))))))).)))))))))..... ( -31.70) >consensus GCUGGACCUGGUCUUGUUCCUGAAUCUGGAU_UUUGACCAUUCACUUUGUUGCCAGCUGUGCGCUGCAUUAGUUGGCCAAGUACAAUGGCCAGAAAAGUGGGUGGAAU .........((((....(((.......))).....))))(((((((((..(((.(((.....))))))....(((((((.......))))))).)))))))))..... (-29.76 = -29.96 + 0.20)



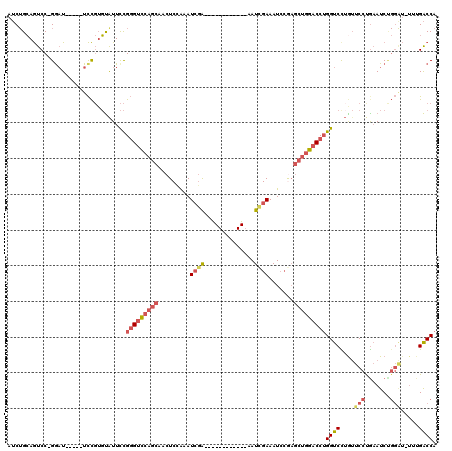
| Location | 17,856,325 – 17,856,438 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.89 |
| Mean single sequence MFE | -32.99 |
| Consensus MFE | -15.83 |
| Energy contribution | -17.19 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17856325 113 - 22224390 AUCUGCAGUCC-GGAU-----UCCGUGUAUUCCGGGUCCAGCAACUCCAAAUCGAAACCGAAACCGAAAUCGAAAUCCAAGCUGGACCUGGUCUUGUUCCUAAAUCUGGAU-UUUGACCA .((...(((((-((((-----(..(..((..(((((((((((.........(((....)))...((....))........)))))))))))...))..)...)))))))))-)..))... ( -36.40) >DroSec_CAF1 113874 102 - 1 AUCUGCAGUCC-GGAU-----UCCGUGUAUUCCGGGUCCAGCAACUCCAAAUCGA------------AAUCGAAAUCCGAGCUGGACCUGGUCCUGUUCCUGAAUCUGGGUUUUUGACCA .((...(..((-((((-----((.(.(.((.(((((((((((...((......))------------..(((.....))))))))))))))....)).)).))))))))..)...))... ( -35.70) >DroSim_CAF1 91000 102 - 1 AUCUGCAGUCC-GGUU-----UCCGUGUAUUCCGGGUCCAGCAACUCCAAAUCGA------------AAUCGAAAUCCGAGCUGGACCUGGUCCUGUUCCUGAAUCUGGGUUUUUGACCA .((...(..((-((.(-----((.(.(.((.(((((((((((...((......))------------..(((.....))))))))))))))....)).)).))).))))..)...))... ( -31.20) >DroEre_CAF1 117589 118 - 1 AUCUGCAGUCC-GAAUCAGAAUCCGUGUGUUCCGGGUCCAGCGACUCCAAAUCAAAAUCGAAAUCGCAAUUGAAAUCCGUGCUGGACCUGGUCUAAUUCCCGAAUCCGGAU-UUUGACCA ...........-...((((((((((..(...((((((((((((........((((...((....))...))))......))))))))))))((........)))..)))))-)))))... ( -37.04) >DroAna_CAF1 127032 87 - 1 UAC-GGAAUCCGGGAU-----UUU--CCAUUUUG-GAAUGGGUACUCUGAAUGGG------------AAUGGAAAUGGAA----UGCCAGGUCCUGAACCUGCAUCA--------GGCCA ...-((..((((....-----(((--((((.(..-((........))..))))))------------))......)))).----..)).((.(((((.......)))--------)))). ( -24.60) >consensus AUCUGCAGUCC_GGAU_____UCCGUGUAUUCCGGGUCCAGCAACUCCAAAUCGA____________AAUCGAAAUCCGAGCUGGACCUGGUCCUGUUCCUGAAUCUGGAU_UUUGACCA .................................(((((((((.........((((..............)))).......)))))))))((((....(((.......))).....)))). (-15.83 = -17.19 + 1.36)

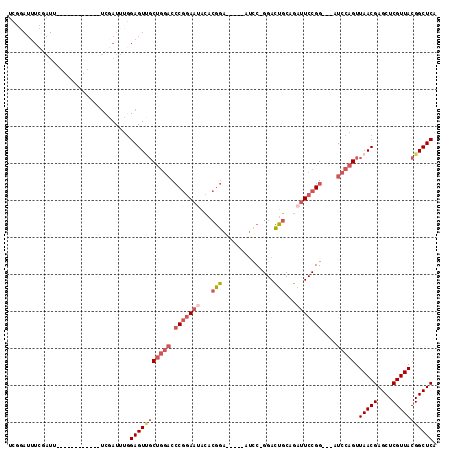
| Location | 17,856,364 – 17,856,471 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.60 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -19.43 |
| Energy contribution | -21.07 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17856364 107 + 22224390 UUGGAUUUCGAUUUCGGUUUCGGUUUCGAUUUGGAGUUGCUGGACCCGGAAUACACGGA-----AUCC-GGACUGCAGAUUCCGG---AUCCAGUUAACGAGCUCGUUACGGCUCA (((((((...(((((((.((((((..(........)..)))))).)))))))...((((-----(((.-(.....).))))))))---)))))).....(((((......))))). ( -37.80) >DroSec_CAF1 113914 95 + 1 UCGGAUUUCGAUU------------UCGAUUUGGAGUUGCUGGACCCGGAAUACACGGA-----AUCC-GGACUGCAGAUUCCGG---AUCCAGUUAACGAGCUCGUUACGGCUCA ((.((..(((...------------.))).)).))(((((((((.(((((((...(((.-----....-...)))...)))))))---.))))).))))(((((......))))). ( -34.60) >DroSim_CAF1 91040 95 + 1 UCGGAUUUCGAUU------------UCGAUUUGGAGUUGCUGGACCCGGAAUACACGGA-----AACC-GGACUGCAGAUUCCGG---AUCCAGUUAACGAGCUCGUUACGGCUCA ((.((..(((...------------.))).)).))(((((((((.(((((((...(((.-----....-...)))...)))))))---.))))).))))(((((......))))). ( -34.60) >DroEre_CAF1 117628 112 + 1 ACGGAUUUCAAUUGCGAUUUCGAUUUUGAUUUGGAGUCGCUGGACCCGGAACACACGGAUUCUGAUUC-GGACUGCAGAUUCCGG---AUCCAGUUAACGAGCUCGUUACGGCUCA .((((((..(((((......)))))..))))))(((((((((((.((((((....(((.(((......-))))))....))))))---.))))))(((((....))))).))))). ( -37.60) >DroYak_CAF1 115643 86 + 1 ---------------------GAUUUCGAUUUGGAGUUGCUGGACCCGGAAUACACGGA-----AUCC-GGACUGCAGAUUCCGG---AUCCAGUUAACGAGCUCGUUACGGCUCA ---------------------..............(((((((((.(((((((...(((.-----....-...)))...)))))))---.))))).))))(((((......))))). ( -31.00) >DroAna_CAF1 127060 95 + 1 UUCCAUUUCCAUU------------CCCAUUCAGAGUACCCAUUC-CAAAAUGG--AAA-----AUCCCGGAUUCC-GUAUCCGGAACAUUCAGUUAACGAGCUCGUUACGGCUCA .............------------........((((..(((((.-...)))))--...-----...((((((...-..))))))...)))).......(((((......))))). ( -21.40) >consensus UCGGAUUUCGAUU____________UCGAUUUGGAGUUGCUGGACCCGGAAUACACGGA_____AUCC_GGACUGCAGAUUCCGG___AUCCAGUUAACGAGCUCGUUACGGCUCA .................................(((((((((((.(((((((...(((..............)))...)))))))....))))).(((((....))))))))))). (-19.43 = -21.07 + 1.64)


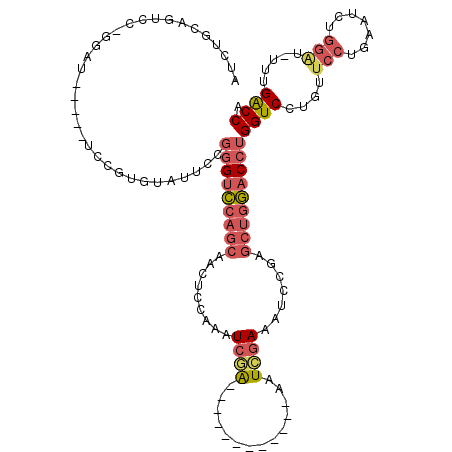
| Location | 17,856,364 – 17,856,471 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.60 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -18.81 |
| Energy contribution | -18.82 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17856364 107 - 22224390 UGAGCCGUAACGAGCUCGUUAACUGGAU---CCGGAAUCUGCAGUCC-GGAU-----UCCGUGUAUUCCGGGUCCAGCAACUCCAAAUCGAAACCGAAACCGAAAUCGAAAUCCAA .((((((...)).))))(((..((((((---(((((((..(((...(-((..-----.))))))))))))))))))).)))......((((...((....))...))))....... ( -34.00) >DroSec_CAF1 113914 95 - 1 UGAGCCGUAACGAGCUCGUUAACUGGAU---CCGGAAUCUGCAGUCC-GGAU-----UCCGUGUAUUCCGGGUCCAGCAACUCCAAAUCGA------------AAUCGAAAUCCGA .((((((...)).))))(((..((((((---(((((((..(((...(-((..-----.))))))))))))))))))).)))......(((.------------...)))....... ( -32.50) >DroSim_CAF1 91040 95 - 1 UGAGCCGUAACGAGCUCGUUAACUGGAU---CCGGAAUCUGCAGUCC-GGUU-----UCCGUGUAUUCCGGGUCCAGCAACUCCAAAUCGA------------AAUCGAAAUCCGA .((((((...)).))))(((..((((((---(((((((..(((...(-((..-----.))))))))))))))))))).)))......(((.------------...)))....... ( -32.50) >DroEre_CAF1 117628 112 - 1 UGAGCCGUAACGAGCUCGUUAACUGGAU---CCGGAAUCUGCAGUCC-GAAUCAGAAUCCGUGUGUUCCGGGUCCAGCGACUCCAAAUCAAAAUCGAAAUCGCAAUUGAAAUCCGU .(((.(((((((....))))).((((((---(((((((..((..((.-......))....))..))))))))))))))).)))....((((...((....))...))))....... ( -32.40) >DroYak_CAF1 115643 86 - 1 UGAGCCGUAACGAGCUCGUUAACUGGAU---CCGGAAUCUGCAGUCC-GGAU-----UCCGUGUAUUCCGGGUCCAGCAACUCCAAAUCGAAAUC--------------------- .((((((...)).))))(((..((((((---(((((((..(((...(-((..-----.))))))))))))))))))).)))..............--------------------- ( -30.60) >DroAna_CAF1 127060 95 - 1 UGAGCCGUAACGAGCUCGUUAACUGAAUGUUCCGGAUAC-GGAAUCCGGGAU-----UUU--CCAUUUUG-GAAUGGGUACUCUGAAUGGG------------AAUGGAAAUGGAA .((((((...)).))))...........(((((((((..-...)))))))))-----(((--(((((((.-(..(((.....)))..).))------------))))))))..... ( -25.90) >consensus UGAGCCGUAACGAGCUCGUUAACUGGAU___CCGGAAUCUGCAGUCC_GGAU_____UCCGUGUAUUCCGGGUCCAGCAACUCCAAAUCGA____________AAUCGAAAUCCGA .((((((...)).)))).....((((((...(((((((..(((.....(((......))).))))))))))))))))....................................... (-18.81 = -18.82 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:27 2006