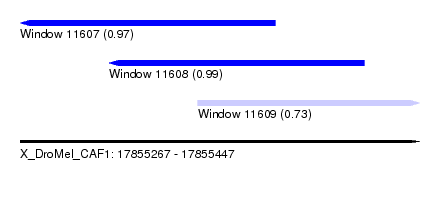
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,855,267 – 17,855,447 |
| Length | 180 |
| Max. P | 0.989887 |

| Location | 17,855,267 – 17,855,382 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -34.44 |
| Consensus MFE | -26.85 |
| Energy contribution | -27.73 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
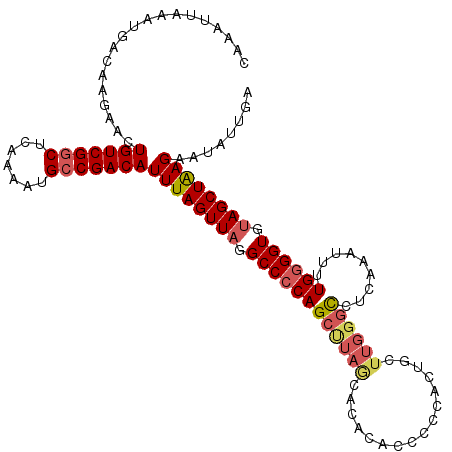
>X_DroMel_CAF1 17855267 115 - 22224390 CAAAUUAAAUGACACGAACUGUCGGCUCAAAAUGCCGACAUUUAGUUAGGCCCCAGCUUAGCACACACGCCCACUGGUUGGGUUUCAAAUUUUGUGGUGUAGCUAAGAAUAUUGA ..............(((..(((((((.......)))))))((((((((.(((.((((...))......(((((.....))))).........)).))).))))))))....))). ( -31.30) >DroSec_CAF1 112844 115 - 1 CAAAUCAAAUGACAAGAACUGUCGGCUCAAAAUGCCGACAUUUAGUUAGGCCCCAGCUUAGCACACACCCGCACUGCUUGGGUCUCAAAUUUUGGGGUGUAGCUGAGAAUAUUGA ....((((...........(((((((.......)))))))((((((((.((((((((...))....(((((.......))))).........)))))).))))))))....)))) ( -38.10) >DroSim_CAF1 89958 115 - 1 CAAAUUAAAUGACAAGAACUGUCGGCUCAAAAUGCCGACAUUUAGUUAGGCCCCAGCUUAGCACACACCCGCACUGCUUGGGCCUCAAAUUUUGGGGUGUAGCUGAGAAUAUUGA ............(((....(((((((.......)))))))((((((((.((((((((((((((...........))).))))).........)))))).))))))))....))). ( -36.70) >DroEre_CAF1 116564 104 - 1 CAAAUUAAAUGACAAGAACUGUCGGCUCAAACUGACGACAUUUAGUUAGGCCCCAGCUUAACACAUACCCC-----------CCACCGAUUUUGGGGUGUAGCUAAGCAUAUGGG ..........((((.....))))((((..((((((......)))))).))))((((((((.....(((.((-----------(((.......))))).)))..)))))...))). ( -28.20) >DroYak_CAF1 114800 115 - 1 CAAAUUAAAUGACAAGAACUGUCGGCUCAAAAUGCCGACAUUUAGUUAGGCCCCAGCCUAACACACACCUCCGCCGCUUGGGCCACUGAAUUUGGGGUGGAGCUAAGCAUAUUGA ............(((....(((((((.......)))))))....(((((((....))))))).............((((((.(((((........)))))..))))))...))). ( -37.90) >consensus CAAAUUAAAUGACAAGAACUGUCGGCUCAAAAUGCCGACAUUUAGUUAGGCCCCAGCUUAGCACACACCCCCACUGCUUGGGCCUCAAAUUUUGGGGUGUAGCUAAGAAUAUUGA ...................(((((((.......)))))))((((((((.((((((((((((................)))))).........)))))).))))))))........ (-26.85 = -27.73 + 0.88)


| Location | 17,855,307 – 17,855,422 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 91.39 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -25.28 |
| Energy contribution | -25.48 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17855307 115 - 22224390 GCUUAAUUUCGAACAAGCCGUGAAUCUUCCCGGCAAAUGACAAAUUAAAUGACACGAACUGUCGGCUCAAAAUGCCGACAUUUAGUUAGGCCCCAGCUUAGCACACACGCCCACU ((.....((((.....((((.((....)).)))).(((.....)))........)))).(((((((.......)))))))....(((((((....)))))))......))..... ( -29.40) >DroSec_CAF1 112884 115 - 1 GCUUAAUUUCGAACAAGCCGUGAAUCUUGCCGGCAAAUGACAAAUCAAAUGACAAGAACUGUCGGCUCAAAAUGCCGACAUUUAGUUAGGCCCCAGCUUAGCACACACCCGCACU ((((..........)))).(((..(((((....((..(((....)))..)).)))))..(((((((.......)))))))....(((((((....))))))).)))......... ( -28.00) >DroSim_CAF1 89998 115 - 1 GCUUAAUUUCGAACAAGCCGUGAAUCUUGCCGGCAAAUGACAAAUUAAAUGACAAGAACUGUCGGCUCAAAAUGCCGACAUUUAGUUAGGCCCCAGCUUAGCACACACCCGCACU ((((..........)))).(((..(((((.((...(((.....)))...)).)))))..(((((((.......)))))))....(((((((....))))))).)))......... ( -28.40) >DroEre_CAF1 116597 111 - 1 GCUUAAUUUCGAACGAGCCGUGGAUCUGCCCGGCAAAUGACAAAUUAAAUGACAAGAACUGUCGGCUCAAACUGACGACAUUUAGUUAGGCCCCAGCUUAACACAUACCCC---- ..(((((((.......((((.((.....)))))).......)))))))(((........((((((......)))))).......(((((((....))))))).))).....---- ( -24.54) >DroYak_CAF1 114840 115 - 1 GCUUAAUUUCGAACGAGUCGUGGAUCUCCACGGCAAAUGACAAAUUAAAUGACAAGAACUGUCGGCUCAAAAUGCCGACAUUUAGUUAGGCCCCAGCCUAACACACACCUCCGCC (((((((((.......(((((((....))))))).......)))))))...........(((((((.......)))))))....(((((((....)))))))..........)). ( -36.64) >consensus GCUUAAUUUCGAACAAGCCGUGAAUCUUCCCGGCAAAUGACAAAUUAAAUGACAAGAACUGUCGGCUCAAAAUGCCGACAUUUAGUUAGGCCCCAGCUUAGCACACACCCCCACU ..(((((((.......((((.((....)).)))).......)))))))...........(((((((.......)))))))....(((((((....)))))))............. (-25.28 = -25.48 + 0.20)


| Location | 17,855,347 – 17,855,447 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.08 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -20.37 |
| Energy contribution | -22.09 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17855347 100 + 22224390 GCAUUUUGAGCCGACAGUUCGUGUCAUUUAAUUUGUCAUUUGCCGGGAAGAUUCACGGCUUGUUCGAAAUUAAGCGACGUCUGUUGGCCGCUGUC-GUU--------------GC (((...(((((((((((....((((.((((((((...((..((((.((....)).))))..))...)))))))).)))).)))))))).....))-).)--------------)) ( -29.60) >DroPse_CAF1 128134 84 + 1 GU---------------UUCUUGUCAUUUAAUUUGUCAUUUGCCAUGGGG--CCAGAGCUGGGGCCAAAUUAAGCGAUGUCUAAUGGCUCUUUUCCUGU--------------GG ..---------------....((.((.......)).))....((((((((--..((((((.((((.............))))...))))))..))))))--------------)) ( -20.72) >DroSec_CAF1 112924 100 + 1 GCAUUUUGAGCCGACAGUUCUUGUCAUUUGAUUUGUCAUUUGCCGGCAAGAUUCACGGCUUGUUCGAAAUUAAGCGACGUCUGUUGGCCGCUGUC-UGU--------------GC ((((..((.((((((((....((((.((((((((...((..((((..........))))..))...)))))))).)))).)))))))))).....-.))--------------)) ( -30.30) >DroSim_CAF1 90038 100 + 1 GCAUUUUGAGCCGACAGUUCUUGUCAUUUAAUUUGUCAUUUGCCGGCAAGAUUCACGGCUUGUUCGAAAUUAAGCGACGUCUGUUGGCCGCUGUC-UGU--------------GC ((((..((.((((((((....((((.((((((((...((..((((..........))))..))...)))))))).)))).)))))))))).....-.))--------------)) ( -30.20) >DroEre_CAF1 116633 93 + 1 UCAGUUUGAGCCGACAGUUCUUGUCAUUUAAUUUGUCAUUUGCCGGGCAGAUCCACGGCUCGUUCGAAAUUAAGCGACGUCUGUUGGCCGU----------------------GC ...((.((.((((((((....((((.((((((((.......((((((.....)).)))).......)))))))).)))).)))))))))).----------------------)) ( -30.54) >DroYak_CAF1 114880 114 + 1 GCAUUUUGAGCCGACAGUUCUUGUCAUUUAAUUUGUCAUUUGCCGUGGAGAUCCACGACUCGUUCGAAAUUAAGCGACGUCUGUUGGCCGCUGUC-UGUGUGUCUGUGUAUCGUC ((((...((((((((((....((((.((((((((.......(.(((((....))))).).......)))))))).)))).))))))))(((....-.)))..)).))))...... ( -32.64) >consensus GCAUUUUGAGCCGACAGUUCUUGUCAUUUAAUUUGUCAUUUGCCGGGAAGAUCCACGGCUUGUUCGAAAUUAAGCGACGUCUGUUGGCCGCUGUC_UGU______________GC .........((((((((....((((.((((((((.......((((.((....)).)))).......)))))))).)))).))))))))........................... (-20.37 = -22.09 + 1.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:21 2006