| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,836,385 – 17,836,510 |
| Length | 125 |
| Max. P | 0.952973 |

| Location | 17,836,385 – 17,836,487 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.10 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -19.77 |
| Energy contribution | -21.50 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17836385 102 + 22224390 AGCGGCUAUGGCUGCGGGCGCUGCAGUGGGCG-------GUGG-------GUGGUGGCAUAUAUUUUGCACAUUAUGCAUGCAGCUGCCGCAAGUAGCAGGCAGGAAAAGCUG-GAA ..(((((.((.((((..((((.(((((..(((-------.((.-------(((((((((.......))).)))))).))))).))))).))..)).)))).)).....)))))-... ( -40.50) >DroSec_CAF1 94034 106 + 1 AGCGGCUACGGCUGCGGGCGGUGCAGUGGGCG---GGUGGUGG-------GUGGUGGCAUAUAUUUUGCACAUUAUGCAUGCAGCUGCCGCAAGUAGCAGGCAGGAAAAGCUG-GAA .((.(((((.((((((.....)))))).((((---(.(((((.-------(((((((((.......))).)))))).))).)).)))))....)))))..))...........-... ( -41.40) >DroSim_CAF1 66637 106 + 1 AGCGGCUACGGCUGCGGGCGGUGCAGUGGGCG---GGUGGUGA-------GUGGUGGCAUAUAUUUUGCACAUUAUGCAUGCAGCUGCCGCAAGUAGCAGGCAGGAAAAGCUG-GAA .((.(((((.((((((.....)))))).((((---(.(((((.-------(((((((((.......))).)))))).))).)).)))))....)))))..))...........-... ( -39.30) >DroEre_CAF1 96017 106 + 1 AGCGGCGAUGGCUGGA----GUGCAAUGGGUGGCGGGUGGUGG-------GUGGUGGCAUAUAUUUUGCACAUUAUGCAUGCAGCUGCCGCAAGUAGCAGGCAGGAAAAGCGGGGAA ..(.((.....((.(.----.(((.((..(((((((.(((((.-------(((((((((.......))).)))))).))).)).)))))))..)).)))..).))....)).).... ( -38.40) >DroYak_CAF1 94644 100 + 1 AGCGCCGAUGGCUGCG----GUGCACUGGGUGGUGGGUGAU------------GUGGCAUAUAUUUUGCACAUUAUGCAUGCAGCUGCCGCAAGUAGCAGGCAGGAAAAGCUG-GAA .((((((.......))----)))).(..(.(.(((.(((((------------(((.((.......)))))))))).)))....(((((((.....)).)))))....).)..-).. ( -36.50) >DroPer_CAF1 111214 103 + 1 GUAGGGGAAGACCUGGAGAGGGACUUUGGGGG---GAUGGCGGGGAUGAGCUGGUGGUGCA-----UUCACAUUAUUCAUGCAACUGCCGCAAGUGGCAGGCAACAUCAGC------ ...........((..(((.....)))..))..---(((((((.((((((....((((....-----.)))).)))))).)))..((((((....))))))....))))...------ ( -33.10) >consensus AGCGGCGAUGGCUGCGGGCGGUGCAGUGGGCG___GGUGGUGG_______GUGGUGGCAUAUAUUUUGCACAUUAUGCAUGCAGCUGCCGCAAGUAGCAGGCAGGAAAAGCUG_GAA .(((((....))))).......((..........................(((((((((.......))).))))))........(((((((.....)).))))).....))...... (-19.77 = -21.50 + 1.73)

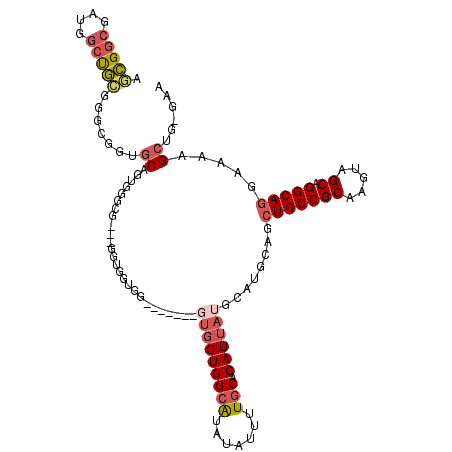

| Location | 17,836,417 – 17,836,510 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 87.51 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -24.24 |
| Energy contribution | -25.64 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17836417 93 + 22224390 -GUGGGUGGUGGCAUAUAUUUUGCACAUUAUGCAUGCAGCUGCCGCAAGUAGCAGGCAGGAAAAGCUG-GAA---------AAGC------AAAGCCAAAGCACCAGCCG -..((.((((((((((............)))))..((..(((((((.....)).))))).....(((.-...---------.)))------...)).....))))).)). ( -31.60) >DroSec_CAF1 94069 94 + 1 GGUGGGUGGUGGCAUAUAUUUUGCACAUUAUGCAUGCAGCUGCCGCAAGUAGCAGGCAGGAAAAGCUG-GAA---------AAGC------AAAGCCAAAGCACCAGCCG (((.((((.((((........((((.....)))).....(((((((.....)).))))).....(((.-...---------.)))------...))))...)))).))). ( -33.30) >DroSim_CAF1 66672 94 + 1 GGUGAGUGGUGGCAUAUAUUUUGCACAUUAUGCAUGCAGCUGCCGCAAGUAGCAGGCAGGAAAAGCUG-GAA---------AAGC------AAAGCCAAAGCACCAGCCG (((..(((.((((........((((.....)))).....(((((((.....)).))))).....(((.-...---------.)))------...))))...)))..))). ( -30.20) >DroEre_CAF1 96051 110 + 1 GGUGGGUGGUGGCAUAUAUUUUGCACAUUAUGCAUGCAGCUGCCGCAAGUAGCAGGCAGGAAAAGCGGGGAAAAGCGGGAAAAGCAAAGCCAAAGCCAAAGCACCAGCCG (((.((((.((((........((((.....))))(((..(((((((.....)).))))).....((........)).......)))........))))...)))).))). ( -35.80) >DroYak_CAF1 94678 104 + 1 GAU-----GUGGCAUAUAUUUUGCACAUUAUGCAUGCAGCUGCCGCAAGUAGCAGGCAGGAAAAGCUG-GAAAAGCUAGAAAAGCAAAGCCAAAGCCAAAGCACCAGCCG (((-----(((.((.......))))))))..........(((((((.....)).))))).....((((-(....(((.....))).........((....)).))))).. ( -28.10) >consensus GGUGGGUGGUGGCAUAUAUUUUGCACAUUAUGCAUGCAGCUGCCGCAAGUAGCAGGCAGGAAAAGCUG_GAA_________AAGC______AAAGCCAAAGCACCAGCCG (((.((((.((((........((((.....)))).....(((((((.....)).)))))...................................))))...)))).))). (-24.24 = -25.64 + 1.40)


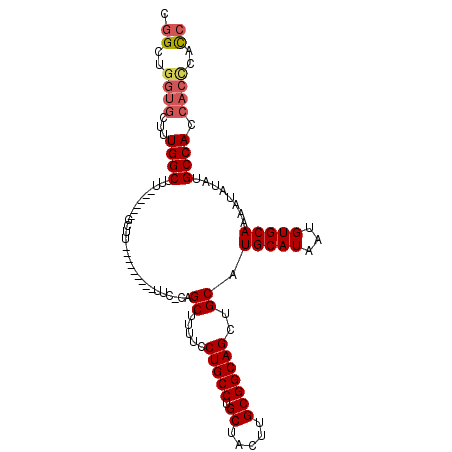
| Location | 17,836,417 – 17,836,510 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 87.51 |
| Mean single sequence MFE | -27.59 |
| Consensus MFE | -22.66 |
| Energy contribution | -23.38 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17836417 93 - 22224390 CGGCUGGUGCUUUGGCUUU------GCUU---------UUC-CAGCUUUUCCUGCCUGCUACUUGCGGCAGCUGCAUGCAUAAUGUGCAAAAUAUAUGCCACCACCCAC- .((.(((((.....((...------(((.---------...-.))).....(((((.((.....)))))))..))..(((((.(((.....))))))))))))).))..- ( -27.10) >DroSec_CAF1 94069 94 - 1 CGGCUGGUGCUUUGGCUUU------GCUU---------UUC-CAGCUUUUCCUGCCUGCUACUUGCGGCAGCUGCAUGCAUAAUGUGCAAAAUAUAUGCCACCACCCACC .((.(((((.....((...------(((.---------...-.))).....(((((.((.....)))))))..))..(((((.(((.....))))))))))))).))... ( -27.10) >DroSim_CAF1 66672 94 - 1 CGGCUGGUGCUUUGGCUUU------GCUU---------UUC-CAGCUUUUCCUGCCUGCUACUUGCGGCAGCUGCAUGCAUAAUGUGCAAAAUAUAUGCCACCACUCACC .((..((((...((((...------(((.---------...-.))).....(((((.((.....))))))).(((((.......)))))........)))).))))..)) ( -25.40) >DroEre_CAF1 96051 110 - 1 CGGCUGGUGCUUUGGCUUUGGCUUUGCUUUUCCCGCUUUUCCCCGCUUUUCCUGCCUGCUACUUGCGGCAGCUGCAUGCAUAAUGUGCAAAAUAUAUGCCACCACCCACC .((.((((((....))...(((..(((.......((........)).....(((((.((.....)))))))..)))(((((...)))))........))))))).))... ( -30.00) >DroYak_CAF1 94678 104 - 1 CGGCUGGUGCUUUGGCUUUGGCUUUGCUUUUCUAGCUUUUC-CAGCUUUUCCUGCCUGCUACUUGCGGCAGCUGCAUGCAUAAUGUGCAAAAUAUAUGCCAC-----AUC ((((((.(((..((((...(((...........((((....-.))))......))).))))...))).))))))...(((((.(((.....))))))))...-----... ( -28.33) >consensus CGGCUGGUGCUUUGGCUUU______GCUU_________UUC_CAGCUUUUCCUGCCUGCUACUUGCGGCAGCUGCAUGCAUAAUGUGCAAAAUAUAUGCCACCACCCACC .((..((((...((((............................((.....(((((.((.....)))))))..)).(((((...)))))........)))).))))..)) (-22.66 = -23.38 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:17 2006