
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,819,481 – 17,819,593 |
| Length | 112 |
| Max. P | 0.995794 |

| Location | 17,819,481 – 17,819,593 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.93 |
| Mean single sequence MFE | -39.67 |
| Consensus MFE | -30.02 |
| Energy contribution | -30.13 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17819481 112 + 22224390 GGCUGGAAGGGGGGGGGUGGGUUGGCAAGAAAAGCCUUUUGGCUUGAUGGGAAUACCCUGCAAGUAGGGGAUAAUCCCAGUGUUAGCUCCACC---UCUGUGGAAGCUAACGAUA ((((......(.(((((((((((((((....(((((....)))))..(((((...(((((....))))).....))))).))))))).)))))---))).)...))))....... ( -46.20) >DroSec_CAF1 77324 98 + 1 GGGGGGA-------GG-------GGCAAGAAAAGCCUUUUGGCUUGAUGGGAAUACCCUGCAAGUAGGGGAUAAUCCCAGUGUUAGCUCCUCC---UCUGUGGAAGCUAACGAUA .((((((-------((-------(((.....(((((....)))))..(((((...(((((....))))).....)))))......))))))))---)))................ ( -42.70) >DroYak_CAF1 77272 96 + 1 GGGG-------------------GGCAAGAAAAGCCUUUUGGCUUGAUGGGAAUACCCUGCAAGUAGGGGAUAAUCCCGGUGUUAGCUCCUUCUCUUCUGUGCAAGCUAACGAUA ((((-------------------(((.....(((((....)))))..(((((...(((((....))))).....)))))......)))))))....................... ( -30.10) >consensus GGGGGGA_______GG_______GGCAAGAAAAGCCUUUUGGCUUGAUGGGAAUACCCUGCAAGUAGGGGAUAAUCCCAGUGUUAGCUCCUCC___UCUGUGGAAGCUAACGAUA ........................((.....(((((....)))))..(((((...(((((....))))).....)))))))(((((((((...........)).))))))).... (-30.02 = -30.13 + 0.11)


| Location | 17,819,481 – 17,819,593 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.93 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -27.73 |
| Energy contribution | -28.07 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
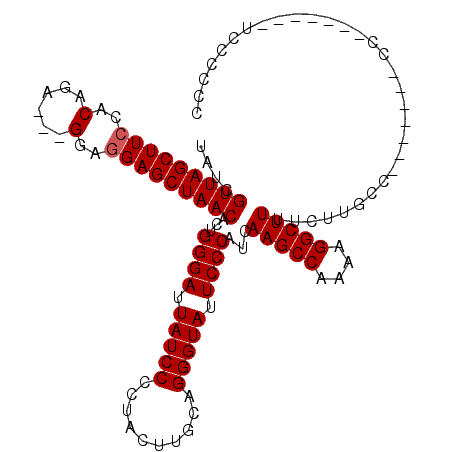
>X_DroMel_CAF1 17819481 112 - 22224390 UAUCGUUAGCUUCCACAGA---GGUGGAGCUAACACUGGGAUUAUCCCCUACUUGCAGGGUAUUCCCAUCAAGCCAAAAGGCUUUUCUUGCCAACCCACCCCCCCCCUUCCAGCC ....(((((((.((((...---.))))))))))).((((((..........((((..(((....)))..))))......(((.......)))...............)))))).. ( -34.20) >DroSec_CAF1 77324 98 - 1 UAUCGUUAGCUUCCACAGA---GGAGGAGCUAACACUGGGAUUAUCCCCUACUUGCAGGGUAUUCCCAUCAAGCCAAAAGGCUUUUCUUGCC-------CC-------UCCCCCC ....((((((((((.....---.).)))))))))..(((((.(((((..........))))).)))))..(((((....)))))........-------..-------....... ( -29.90) >DroYak_CAF1 77272 96 - 1 UAUCGUUAGCUUGCACAGAAGAGAAGGAGCUAACACCGGGAUUAUCCCCUACUUGCAGGGUAUUCCCAUCAAGCCAAAAGGCUUUUCUUGCC-------------------CCCC ....((((((((.(........)...))))))))...((((.(((((..........))))).))))...(((((....)))))........-------------------.... ( -26.20) >consensus UAUCGUUAGCUUCCACAGA___GGAGGAGCUAACACUGGGAUUAUCCCCUACUUGCAGGGUAUUCCCAUCAAGCCAAAAGGCUUUUCUUGCC_______CC_______UCCCCCC ....(((((((((..(......)..)))))))))...((((.(((((..........))))).))))...(((((....)))))............................... (-27.73 = -28.07 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:12 2006