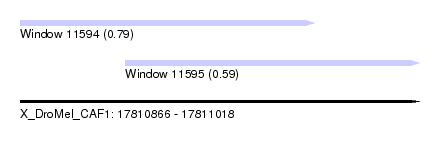
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,810,866 – 17,811,018 |
| Length | 152 |
| Max. P | 0.790576 |

| Location | 17,810,866 – 17,810,978 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.52 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -16.34 |
| Energy contribution | -16.78 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17810866 112 + 22224390 AGCUCGGCCAACAAGUGUAUUACAUUUGCCUCAAAUUUCGAUAUGGAACAAUGCUUAUAUAAAUAUCGCUCAAAUGCCAGCGAUAUUUUUA--GCCCAUCGAUAGCUGGACUUU------ ((..((((...((((((.....)))))).........((((..(((......(((.....((((((((((........))))))))))..)--)))))))))..))))..))..------ ( -27.40) >DroSec_CAF1 68878 111 + 1 AGCUCGGCCAACAAGUGUUUUGCCAUUGCCCCA-CUUUCGAUGUAGAACAAUGCUUAUAUAAAUAUCGCUCACACGCCAGCGAUAUUUUUA--GCCUAUCGAUAGCUGGGCUUU------ ((((((((.......((((((((.((((.....-....))))))))))))..(((.....((((((((((........))))))))))..)--)).........))))))))..------ ( -32.00) >DroEre_CAF1 69229 107 + 1 ---UCGGCCAACAAGAGUUUUGCAAUAGCC-CA-CUUUGAAUAUAGAACAAUGCUUAUAUAAAUAUCGCUAGCACGACAGCGAUGUUUUUA--GGCUAUCGAUAGCUGGGCUUA------ ---...((.(((....)))..))...((((-((-((((((............(((((...((((((((((........)))))))))).))--)))..)))).)).))))))..------ ( -26.94) >DroYak_CAF1 68837 102 + 1 --CUCGGCCAACAAGUGUUUGGCUC----------------UAUGGAACAAUGCUUAUAUAAAUAUCGCUCACACGCAAGUGAUAUUUUUAUAUCCUAUCGAUAGCUGGGCUUAGGCUUG --...((((.......(((..(((.----------------.((.((.....(..((((.((((((((((........)))))))))).))))..)..)).)))))..)))...)))).. ( -23.60) >consensus __CUCGGCCAACAAGUGUUUUGCAAUUGCC_CA_CUUUCGAUAUAGAACAAUGCUUAUAUAAAUAUCGCUCACACGCCAGCGAUAUUUUUA__GCCUAUCGAUAGCUGGGCUUA______ .(((((((................................((((((........))))))((((((((((........))))))))))................)))))))......... (-16.34 = -16.78 + 0.44)

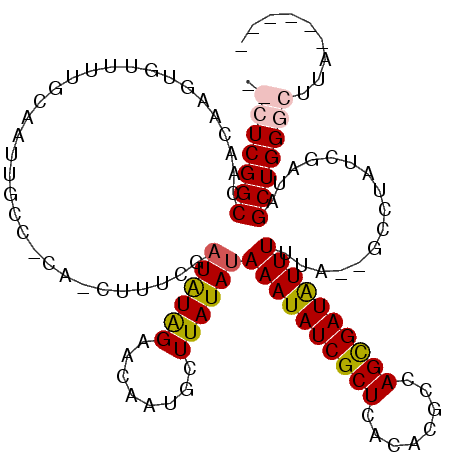
| Location | 17,810,906 – 17,811,018 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.78 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -22.17 |
| Energy contribution | -21.16 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17810906 112 + 22224390 AUAUGGAACAAUGCUUAUAUAAAUAUCGCUCAAAUGCCAGCGAUAUUUUUA--GCCCAUCGAUAGCUGGACUUU------UGUAACUGGCAAGUGAUAGCGAAUCUUUGGCUAAGAUGGC ............(((.....((((((((((........))))))))))..)--))(((((..((((..((..((------(((.(((....)))....)))))..))..)))).))))). ( -33.30) >DroSec_CAF1 68917 112 + 1 AUGUAGAACAAUGCUUAUAUAAAUAUCGCUCACACGCCAGCGAUAUUUUUA--GCCUAUCGAUAGCUGGGCUUU------UGUAACUGGCAAAUGAUAGCGAAUCUGUGGCUAAGAUGGC ............(((.......((((((((........))))))))(((((--(((.((.(((.((((..(.((------(((.....))))).).))))..))).)))))))))).))) ( -28.00) >DroEre_CAF1 69264 112 + 1 AUAUAGAACAAUGCUUAUAUAAAUAUCGCUAGCACGACAGCGAUGUUUUUA--GGCUAUCGAUAGCUGGGCUUA------UGUAACUGGUAUGUGAUAGCGAAUCUCUGGCUAAGAUGGC ............(((((...((((((((((........)))))))))).))--)))((((..((((..(((...------.))....(((.(((....))).))).)..)))).)))).. ( -28.10) >DroYak_CAF1 68860 119 + 1 -UAUGGAACAAUGCUUAUAUAAAUAUCGCUCACACGCAAGUGAUAUUUUUAUAUCCUAUCGAUAGCUGGGCUUAGGCUUGUGUAACUGGUAAGUGAUAGCGAAUCUCUGGCUAAGAUGGU -..............((((.((((((((((........)))))))))).))))..(((((..((((..(....((..((((...(((....)))....))))..)))..)))).))))). ( -29.70) >consensus AUAUAGAACAAUGCUUAUAUAAAUAUCGCUCACACGCCAGCGAUAUUUUUA__GCCUAUCGAUAGCUGGGCUUA______UGUAACUGGCAAGUGAUAGCGAAUCUCUGGCUAAGAUGGC ((((((........))))))((((((((((........)))))))))).......(((((....((..(................)..))......((((.(.....).)))).))))). (-22.17 = -21.16 + -1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:09 2006