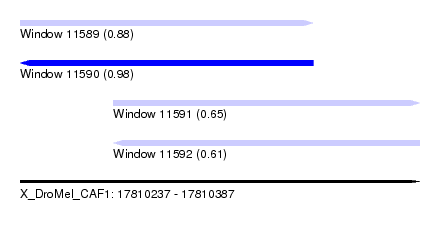
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,810,237 – 17,810,387 |
| Length | 150 |
| Max. P | 0.980769 |

| Location | 17,810,237 – 17,810,347 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -22.34 |
| Energy contribution | -22.57 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17810237 110 + 22224390 AAUUUAAUGUUCCUAAGGAUAAUGUUUAAUUUGCUUUUAU--UUUCGCAUACCCGUAACCAUCGAGGGAGUUGCGCUGGAGCUGCUGCUCCCGCAGCUGCAGCUUCCGCUCG .......(((((....)))))...................--....((.....((.......)).(((((((((((((((((....)))))...))).)))))))))))... ( -29.60) >DroPse_CAF1 79382 88 + 1 -------------UAAGGCUAGAGUUAAGG------UUA-----AGGCAUACCCGUAACCAUCGAGCGAGUUGCGCUGGAGCUGCUGUUCGCGGAGCUGCAGCUUUCGCUCG -------------...............((------(((-----.((.....)).)))))..((((((((....((((.((((.(((....))))))).)))).)))))))) ( -34.30) >DroSec_CAF1 68269 110 + 1 AAUUCAAGGUUCUUGAAGAUAAUGUUUAAUUUGCUUUGAU--UUUUGCAUACCCGUAACCAUCGAGGGAGUUGCGCUGUAGCUGCUGCUCCCGCAGCUGCAGCUUCCGCUCG ..((((((...))))))................(((((((--..((((......))))..)))))))((((.(.(((((((((((.......)))))))))))..).)))). ( -36.80) >DroEre_CAF1 68593 112 + 1 AAUUCAAGGUUCUUGAGGAUAAUGUUUAAUUUGUUUGUUUUGCUUGGCAUACCCGUAACCGUCGAGGGAAUUGCGCUGGAGCUGCUGCUCCCGCAGCUGCAACUUCCGCUCG .......((((..((.((...(((((......((.......))..))))).)))).))))..((((((((((((((((((((....)))))...))).)))).)))).)))) ( -31.10) >DroYak_CAF1 68217 105 + 1 ---AGCGGCUUUUUGGGAAUAAUGUUUAAUUG----AUUUUGCUUGGCAUACCCGUAACCGUCGAGGGAGUUGCGCUGGAGCUGCUGCUCCCGCAGCUGCAGCUUCCGCUCG ---((((((....((((..((((.....))))----....(((...)))..)))).....)))..(((((((((((((((((....)))))...))).)))))))))))).. ( -36.80) >DroPer_CAF1 80589 88 + 1 -------------UAAGGCUAGAGUUAAGG------UUA-----AGGCAUACCCGUAACCAUCGAGCGAGUUGCGCUGGAGCUGCUGUUCGCGGAGCUGCAGCUUUCGCUCG -------------...............((------(((-----.((.....)).)))))..((((((((....((((.((((.(((....))))))).)))).)))))))) ( -34.30) >consensus ___U_AAGGUUCUUAAGGAUAAUGUUUAAUUU____UUAU___UUGGCAUACCCGUAACCAUCGAGGGAGUUGCGCUGGAGCUGCUGCUCCCGCAGCUGCAGCUUCCGCUCG ..............................................(((..(((...........)))...)))((.((((((((.(((.....))).)))))))).))... (-22.34 = -22.57 + 0.22)



| Location | 17,810,237 – 17,810,347 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -32.07 |
| Consensus MFE | -20.09 |
| Energy contribution | -19.53 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17810237 110 - 22224390 CGAGCGGAAGCUGCAGCUGCGGGAGCAGCAGCUCCAGCGCAACUCCCUCGAUGGUUACGGGUAUGCGAAA--AUAAAAGCAAAUUAAACAUUAUCCUUAGGAACAUUAAAUU .(((.(((.(.(((.(((((....))))).((....))))).)))))))((((((....(((.(((....--......))).)))....))))))................. ( -30.50) >DroPse_CAF1 79382 88 - 1 CGAGCGAAAGCUGCAGCUCCGCGAACAGCAGCUCCAGCGCAACUCGCUCGAUGGUUACGGGUAUGCCU-----UAA------CCUUAACUCUAGCCUUA------------- (((((((..((((.((((..((.....)))))).)))).....)))))))..(((((.(((....)))-----)))------))...............------------- ( -31.40) >DroSec_CAF1 68269 110 - 1 CGAGCGGAAGCUGCAGCUGCGGGAGCAGCAGCUACAGCGCAACUCCCUCGAUGGUUACGGGUAUGCAAAA--AUCAAAGCAAAUUAAACAUUAUCUUCAAGAACCUUGAAUU .(((.(((.(.(((.(((((....))))).((....))))).)))))))((((((....(((.(((....--......))).)))....))))))((((((...)))))).. ( -33.10) >DroEre_CAF1 68593 112 - 1 CGAGCGGAAGUUGCAGCUGCGGGAGCAGCAGCUCCAGCGCAAUUCCCUCGACGGUUACGGGUAUGCCAAGCAAAACAAACAAAUUAAACAUUAUCCUCAAGAACCUUGAAUU ((((.((((.((((.(((((....))))).((....))))))))))))))..((((..(((((............................))))).....))))....... ( -33.19) >DroYak_CAF1 68217 105 - 1 CGAGCGGAAGCUGCAGCUGCGGGAGCAGCAGCUCCAGCGCAACUCCCUCGACGGUUACGGGUAUGCCAAGCAAAAU----CAAUUAAACAUUAUUCCCAAAAAGCCGCU--- ((((.(((.(.(((.(((((....))))).((....))))).)))))))).(((((..(((.(((...........----........)))....)))....)))))..--- ( -32.81) >DroPer_CAF1 80589 88 - 1 CGAGCGAAAGCUGCAGCUCCGCGAACAGCAGCUCCAGCGCAACUCGCUCGAUGGUUACGGGUAUGCCU-----UAA------CCUUAACUCUAGCCUUA------------- (((((((..((((.((((..((.....)))))).)))).....)))))))..(((((.(((....)))-----)))------))...............------------- ( -31.40) >consensus CGAGCGGAAGCUGCAGCUGCGGGAGCAGCAGCUCCAGCGCAACUCCCUCGAUGGUUACGGGUAUGCCAA___AUAA____AAAUUAAACAUUAUCCUCAAGAACCUU_A___ ..(((....)))((((((..(((((..((.((....))))..)))))....((....))))).))).............................................. (-20.09 = -19.53 + -0.55)


| Location | 17,810,272 – 17,810,387 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 83.90 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -27.35 |
| Energy contribution | -26.88 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17810272 115 + 22224390 UUUAU--UUUCGCAUACCCGUAACCAUCGAGGGAGUUGCGCUGGAGCUGCUGCUCCCGCAGCUGCAGCUUCCGCUCGGCGAUCUUCUUUUGCUCCUCCUUCUGCUCGAGCUGCUCCU .....--....(((..............(((((((..((((.(((((....))))).)).((((.(((....)))))))...........))..))))))).((....))))).... ( -39.40) >DroGri_CAF1 84729 112 + 1 -----UGGCAAGCAUACCCGUAGCCAUCGAGUGAGUUGCGCUGUUGCUGCUGCUCUCUGAGCUGCAAUUUGCGCUCUGCGAUCUUCUUUUGCUCCUCCUUCUGCUCGAGCUGCUCCU -----..............(((((..(((((((((..((((.(((((....((((...)))).)))))..))))...((((.......))))..))).....))))))))))).... ( -34.30) >DroSec_CAF1 68304 115 + 1 UUGAU--UUUUGCAUACCCGUAACCAUCGAGGGAGUUGCGCUGUAGCUGCUGCUCCCGCAGCUGCAGCUUCCGCUCGGCGAUCUUCUUUUGCUCCUCCUUCUGCUCGAGCUGCUCCU (((((--..((((......))))..)))))((((((...(((((((((((.......)))))))))))....(((((((((..................)).)).))))).)))))) ( -42.77) >DroEre_CAF1 68628 117 + 1 UGUUUUGCUUGGCAUACCCGUAACCGUCGAGGGAAUUGCGCUGGAGCUGCUGCUCCCGCAGCUGCAACUUCCGCUCGGCGAUCUUCUUCUGCUCCUCCUUCUGCUCGAGCUGCUCCU .((...((((((((((....))..(((((((((((((((((((((((....)))))...))).)))).)))).))))))).....................))).))))).)).... ( -40.80) >DroMoj_CAF1 100078 111 + 1 ------GUGAAGCAUACCCAUAGCCAUCGAGAGAGUUGCGCUGCUGCUGCUGCUCUCGUAGCUGCAAUUUGCGCUCGGCAAUCUUCUUCUGCUCCUCCUUCUGCUCGAGCUGUUCCU ------.............(((((..(((((((((..((((...(((.(((((....))))).)))....))))..((((.........)))).....)))).)))))))))).... ( -32.30) >DroAna_CAF1 77422 112 + 1 CUGA-----UGUCAUACCCAUAUCCAUCGAGGGAGUUGCGCUGCAGUUGCUGCUCUCGGAGUUGCAACUUCCGUUCGGCGAUCUUCUUCUGCUCCUCCUUCUGUUCGAGUUGUUCCU ..((-----((.............))))(((((((..((((((.((((((.((((...)))).))))))......))))((......)).))..)))))))................ ( -30.32) >consensus UU_A___UUUAGCAUACCCGUAACCAUCGAGGGAGUUGCGCUGCAGCUGCUGCUCCCGCAGCUGCAACUUCCGCUCGGCGAUCUUCUUCUGCUCCUCCUUCUGCUCGAGCUGCUCCU ...........(((................(((((((((((....)).(((((....))))).)))))))))((((((((.....................))).)))))))).... (-27.35 = -26.88 + -0.47)


| Location | 17,810,272 – 17,810,387 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.90 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -29.09 |
| Energy contribution | -29.48 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17810272 115 - 22224390 AGGAGCAGCUCGAGCAGAAGGAGGAGCAAAAGAAGAUCGCCGAGCGGAAGCUGCAGCUGCGGGAGCAGCAGCUCCAGCGCAACUCCCUCGAUGGUUACGGGUAUGCGAAA--AUAAA ....(((((((((((.((.((((..((.........((((...))))..((((.((((((.......)))))).))))))..)))).))....))).))))).)))....--..... ( -43.50) >DroGri_CAF1 84729 112 - 1 AGGAGCAGCUCGAGCAGAAGGAGGAGCAAAAGAAGAUCGCAGAGCGCAAAUUGCAGCUCAGAGAGCAGCAGCAACAGCGCAACUCACUCGAUGGCUACGGGUAUGCUUGCCA----- ..(((((((((((((.((..(((..((...........))...((((...((((.((((...)))).)))).....))))..)))..))....))).))))).)))))....----- ( -35.50) >DroSec_CAF1 68304 115 - 1 AGGAGCAGCUCGAGCAGAAGGAGGAGCAAAAGAAGAUCGCCGAGCGGAAGCUGCAGCUGCGGGAGCAGCAGCUACAGCGCAACUCCCUCGAUGGUUACGGGUAUGCAAAA--AUCAA ....(((((((((((.((.((((..((.........((((...))))..((((.((((((.......)))))).))))))..)))).))....))).))))).)))....--..... ( -43.50) >DroEre_CAF1 68628 117 - 1 AGGAGCAGCUCGAGCAGAAGGAGGAGCAGAAGAAGAUCGCCGAGCGGAAGUUGCAGCUGCGGGAGCAGCAGCUCCAGCGCAAUUCCCUCGACGGUUACGGGUAUGCCAAGCAAAACA ..(.((((((((.((..........)).......(((((.((((.((((.((((.(((((....))))).((....)))))))))))))).))))).))))).)))).......... ( -42.50) >DroMoj_CAF1 100078 111 - 1 AGGAACAGCUCGAGCAGAAGGAGGAGCAGAAGAAGAUUGCCGAGCGCAAAUUGCAGCUACGAGAGCAGCAGCAGCAGCGCAACUCUCUCGAUGGCUAUGGGUAUGCUUCAC------ .(((.((((((((((.((..((((.((((.......)))))..((((...((((.(((.(....).))).))))..))))..)))..))....))).))))).)).)))..------ ( -34.10) >DroAna_CAF1 77422 112 - 1 AGGAACAACUCGAACAGAAGGAGGAGCAGAAGAAGAUCGCCGAACGGAAGUUGCAACUCCGAGAGCAGCAACUGCAGCGCAACUCCCUCGAUGGAUAUGGGUAUGACA-----UCAG ................((.((((..((............((....)).((((((..(((...)))..)))))).....))..)))).))((((.(((....)))..))-----)).. ( -25.40) >consensus AGGAGCAGCUCGAGCAGAAGGAGGAGCAAAAGAAGAUCGCCGAGCGGAAGUUGCAGCUGCGAGAGCAGCAGCUACAGCGCAACUCCCUCGAUGGUUACGGGUAUGCCAAA___U_AA ....(((((((((((.((.((((..((....(.....).....((...((((((.(((.....))).))))))...))))..)))).))....))).))))).)))........... (-29.09 = -29.48 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:30:06 2006