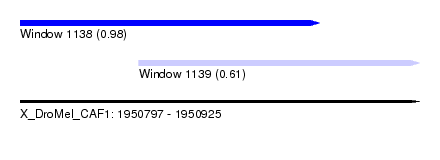
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,950,797 – 1,950,925 |
| Length | 128 |
| Max. P | 0.975773 |

| Location | 1,950,797 – 1,950,893 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -47.37 |
| Consensus MFE | -31.16 |
| Energy contribution | -32.50 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1950797 96 + 22224390 UUGGUCGC-GGGGGGGCGCCAGGGUGGAUGCAGCGGAUCCGUUGCCUCU------GCACUGCUUCAUCCUCAGCCUCAGCCUGUUGCCCACCGCGGAAGCAGU ....((((-((..(((((.(((((..((.((((((....)))))).)).------.)...(((........))).....)))).))))).))))))....... ( -44.30) >DroSec_CAF1 13269 96 + 1 UUGGGCGC-GGGGGGGCGUCAGGGUGAAUGCAGCGGAUCUGUUGCCGCU------GCACUGCUUCAUCCUCAGCCUCAGCCUGUUGCCCACCGCGGAAGCAGU .(((((((-((((((((...((((((((((((((((........)))))------)))....))))))))..)))))..)))).))))))..((....))... ( -53.40) >DroEre_CAF1 14478 91 + 1 UGGGGCGAAGGGGGGGCGCAAGGGUGGAUGCAGCGGAGCCGUUGCCAUUACCGUUGCACUGCUU------C------AGCCUGUUGCCCACCGCGUAAGCAGU ..(((((((((.(((((((((.(((((..((((((....))))))..))))).))))...))))------)------..))).))))))...((....))... ( -44.40) >consensus UUGGGCGC_GGGGGGGCGCCAGGGUGGAUGCAGCGGAUCCGUUGCCACU______GCACUGCUUCAUCCUCAGCCUCAGCCUGUUGCCCACCGCGGAAGCAGU .........((..(((((.((((.(((..((((((....))))))..........((...)).............))).)))).))))).))((....))... (-31.16 = -32.50 + 1.34)

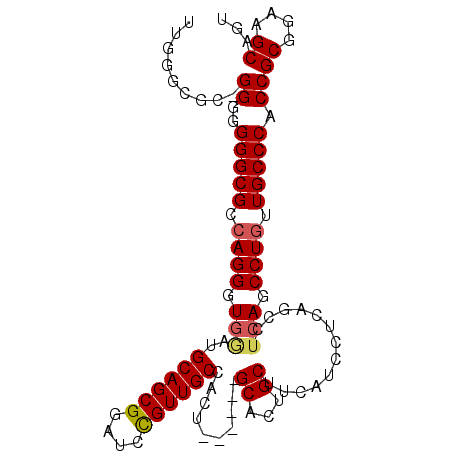
| Location | 1,950,835 – 1,950,925 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.21 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -20.27 |
| Energy contribution | -21.27 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1950835 90 + 22224390 CGUUGCCUCU------GCACUGCUUCAUCCUCAGCCUCAGCCUGUUGCCCACCGCGGAAGCAGUGCGGUAACCAGUGACAAUUG-ACCGA--AAACGAU .((((((...------((((((((((.....((((....).))).(((.....)))))))))))))))))))((((....))))-.....--....... ( -27.40) >DroSec_CAF1 13307 90 + 1 UGUUGCCGCU------GCACUGCUUCAUCCUCAGCCUCAGCCUGUUGCCCACCGCGGAAGCAGUGCGGUAACCAGUGACAAUUG-ACCGA--AAACGAU (((..(.(((------((((((((((.....((((....).))).(((.....)))))))))))))))).....)..)))....-.....--....... ( -28.10) >DroEre_CAF1 14517 87 + 1 CGUUGCCAUUACCGUUGCACUGCUU------C------AGCCUGUUGCCCACCGCGUAAGCAGUGCGGUAACCAGUGACAAUUGCACCAGCAAAAAGAU .((..(..(((((...(((((((((------.------.((..((.....)).))..))))))))))))))...)..))..((((....))))...... ( -27.00) >consensus CGUUGCCACU______GCACUGCUUCAUCCUCAGCCUCAGCCUGUUGCCCACCGCGGAAGCAGUGCGGUAACCAGUGACAAUUG_ACCGA__AAACGAU .((((((.........((((((((((.....(((.......))).(((.....)))))))))))))))))))((((....))))............... (-20.27 = -21.27 + 1.00)

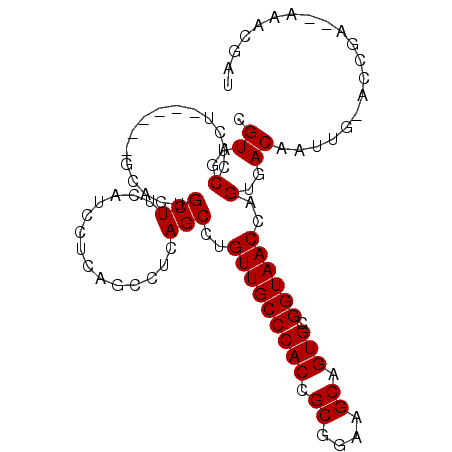
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:51:44 2006