
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,775,801 – 17,775,975 |
| Length | 174 |
| Max. P | 0.980420 |

| Location | 17,775,801 – 17,775,902 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.00 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -3.40 |
| Energy contribution | -3.44 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.14 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17775801 101 + 22224390 AG-AUUACAUUGUGGAUGUUUAUUGA-------GGGGGGAGUGGAAAAAAUUCUCAUUUUGUAAUGCUUCCA----------GCUCAAUCGAAAGGUUAACAACUUACAACUU-UAACAU ..-......((((((.((((....((-------(..(((((((...(((((....)))))....))))))).----------.)))((((....)))))))).).)))))...-...... ( -21.00) >DroSec_CAF1 34558 100 + 1 AG-AUAAAAUUCUGGAUGUUUAUUGA-------UGAGGGAGUGGAAA-CGUCCUCAUUUUAUAAUGCAUCCA----------GCUCAAUCUAAAGGUUAACAACUUACAACUU-UAACAU ((-((......((((((((((((.((-------(((((..((....)-)..)))))))..)))).)))))))----------)....)))).((((((..........)))))-)..... ( -28.70) >DroSim_CAF1 35924 100 + 1 AG-AUAAAAUUCUGGAUGUUUAUUGA-------UGAGGGAGUGGAAA-CAUCCUCAUUUUAUAAUGCAUCCA----------GCUCAAUCUAAAGGUUAACAACUUACAACUU-UAACAU ((-((......((((((((((((.((-------(((((..(((....-))))))))))..)))).)))))))----------)....)))).((((((..........)))))-)..... ( -27.90) >DroEre_CAF1 32786 99 + 1 AU-AUCACAUUCUGGAUGCUUAUUGAGUCAACAGGGGGGAUUGGAAAACAUCCUCGUCUUAUAAUGUUCCCA-------------CAAUCCCUAGGAUAAAAACUUAAAAC-------AU .(-(((....((((...((((...))))...))))(((((((((..(((((............)))))..).-------------))))))))..))))............-------.. ( -20.50) >DroYak_CAF1 33372 113 + 1 AUGAUCACAUUCUGGCUGCUCAGUGA-------GGUGCGAUUGGAAAAAAUCCGCAAUUCAUAAUGUUCCAAUAUGUAACAAGGUCAAUCUGAAGGCUAACUACUUAAGACUUUUAUCAU .(((((((((..(((..((...((((-------(.(((((((......))).)))).)))))...)).)))..)))).....))))).(((.(((........))).))).......... ( -23.10) >consensus AG_AUAACAUUCUGGAUGUUUAUUGA_______GGAGGGAGUGGAAAACAUCCUCAUUUUAUAAUGCUUCCA__________GCUCAAUCUAAAGGUUAACAACUUACAACUU_UAACAU ............((((.((....(((........(((((.((.....)).)))))...)))....)).))))................................................ ( -3.40 = -3.44 + 0.04)


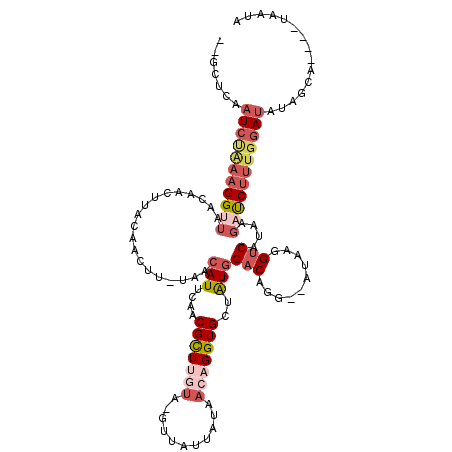
| Location | 17,775,865 – 17,775,975 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.56 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -7.14 |
| Energy contribution | -9.46 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17775865 110 + 22224390 --GCUCAAUCGAAAGGUUAACAACUUACAACUU-UAACAUUCAACGCUUGUA-GUUAUUUAAACAGGUGCUAUGGACAUG--AUAAGGUCAUAAGAUCUUUGGAUAUAGCA----UAAUA --(((..(((.((((((......((((..((((-((.(((((..(((((((.-.........))))))).....)).)))--.))))))..)))))))))).)))..))).----..... ( -23.70) >DroSec_CAF1 34621 114 + 1 --GCUCAAUCUAAAGGUUAACAACUUACAACUU-UAACAUUCAACGCUUGUA-GUCAUUAUAACAGGUGCUAUGGACAGG--AUAAGGUCAUAAGAUCUUUGGAUAUAGCAAAUUUAAUA --(((..((((((((((......((((..((((-((.(.((((.(((((((.-((....)).)))))))...))))...)--.))))))..))))))))))))))..))).......... ( -25.00) >DroSim_CAF1 35987 114 + 1 --GCUCAAUCUAAAGGUUAACAACUUACAACUU-UAACAUUCAACGCUUGUA-GUUAUUAUAACAGGUGCUAUGGACAGG--AUAAGGUCAUAAGAUCUUUGGAUAUAGCAAAUUUAAUA --(((..((((((((((......((((..((((-((.(.((((.(((((((.-((....)).)))))))...))))...)--.))))))..))))))))))))))..))).......... ( -25.80) >DroEre_CAF1 32857 91 + 1 -----CAAUCCCUAGGAUAAAAACUUAAAAC-------AUUCGACGUUUAUA-GUUAUAAUAACCGGUGCUAUGGACAGG--AUUACGACAUA-UACCCUUGGAAUA------------- -----...(((..(((...............-------.......(((((((-((.............))))))))).(.--....)......-...))).)))...------------- ( -13.12) >DroYak_CAF1 33445 103 + 1 AAGGUCAAUCUGAAGGCUAACUACUUAAGACUUUUAUCAUACAACGUUUGUAUGGUAUAAUA---GGUGCUGUGGACAGGUCAUUAGGUCAUA-GACCUUUGUAUAA------------- ((((((.((((((.((((..((((...((((((.(((((((((.....)))))))))....)---))).))))))...)))).))))))....-)))))).......------------- ( -27.40) >consensus __GCUCAAUCUAAAGGUUAACAACUUACAACUU_UAACAUUCAACGCUUGUA_GUUAUUAUAACAGGUGCUAUGGACAGG__AUAAGGUCAUAAGAUCUUUGGAUAUAGCA____UAAUA .......(((((((((((...................(((....(((((((...........)))))))..)))(((..........)))....)))))))))))............... ( -7.14 = -9.46 + 2.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:51 2006