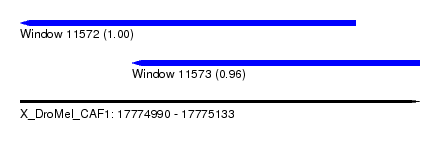
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,774,990 – 17,775,133 |
| Length | 143 |
| Max. P | 0.999993 |

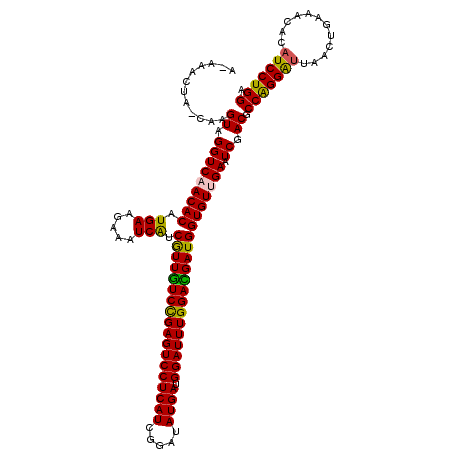
| Location | 17,774,990 – 17,775,110 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.18 |
| Mean single sequence MFE | -39.22 |
| Consensus MFE | -39.52 |
| Energy contribution | -38.72 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.17 |
| Structure conservation index | 1.01 |
| SVM decision value | 5.76 |
| SVM RNA-class probability | 0.999993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17774990 120 - 22224390 ACAAACCACCAAGUAGGUCAACACAUGAAGAAAUCAUCAUUGUCUGAGUCCUCAUCGGAUAUGAUGGAUUUGGACGAUGGUGUCGAGUCCACGCCGGGAUUGACAGAAACACAUCCUGGA ............((.((((.((((((((.....))))(((((((..(((((((((.....)))).)))))..))))))))))).))..)))).(((((((((.......)).))))))). ( -38.20) >DroVir_CAF1 49288 118 - 1 A-AAACCA-CAAGUAGGUCAACACUUGAAGAAAUCAUCGUUGUCCGAGUCCUCAUCGGAUAUGAUGGAUUUGGACGAUGGUGUUGAAUCGACGCCAGGGUUAACUGAAACACAUCCUGGA .-......-...((...(((((((.(((.....))).((((((((((((((((((.....)))).)))))))))))))))))))))....)).(((((((............))))))). ( -42.10) >DroPse_CAF1 44002 120 - 1 ACAAACUACCAAGUAGGUCAACACAUGAAGAAAUCGUCGUUGUCCGAGUCCUCAUCGGAUAUGAUGGAUUUGGAUGAUGGUGUCGAGUCGACGCCGGGAUUAACUGACACACAUCCUGGA .....((((...)))).((.((((((((.....))))(((..(((((((((((((.....)))).)))))))))..))))))).)).......(((((((............))))))). ( -36.30) >DroGri_CAF1 42647 118 - 1 A-AAACUA-CAAGUAGGUCAACACUUGAAGAAAUCAUCGUUGUCUGAGUCCUCAUCGGAUAUGAUGGAUUUGGACGAUGGUGUUGAAUCGACGCCGGGAUUAACUGAAACACAUCCUGGA .-......-...((...(((((((.(((.....))).(((((((..(((((((((.....)))).)))))..))))))))))))))....)).(((((((............))))))). ( -39.50) >DroWil_CAF1 48565 119 - 1 A-ACACUACCAAGUAGGUCAACACAUGAAGAAAUCAUCGUUAUCUGAGUCCUCAUCGGAUAUGAUGGAUUUGGAUGAUGGUGUUGAAUCGACACCAGGACUAACUGAAACGCAUCCUGGA .-...((((...)))).(((((((((((.....))))(((((((..(((((((((.....)))).)))))..)))))))))))))).......((((((..............)))))). ( -38.94) >DroMoj_CAF1 42267 118 - 1 A-AAACUA-CAAGUAGGUCAACACUUGAAGAAAUCAUCGUUGUCCGAGUCCUCAUCGGAUAUGAUGGAUUUGGAUGAUGGUGUUGAAUCGACGCCAGGAUUAACUGAAACGCAUCCUGGA .-......-...((...(((((((.(((.....))).(((..(((((((((((((.....)))).)))))))))..))))))))))....)).(((((((............))))))). ( -40.30) >consensus A_AAACUA_CAAGUAGGUCAACACAUGAAGAAAUCAUCGUUGUCCGAGUCCUCAUCGGAUAUGAUGGAUUUGGACGAUGGUGUUGAAUCGACGCCAGGAUUAACUGAAACACAUCCUGGA ............((.(((((((((.(((.....))).((((((((((((((((((.....)))).))))))))))))))))))))).)).)).(((((((............))))))). (-39.52 = -38.72 + -0.80)


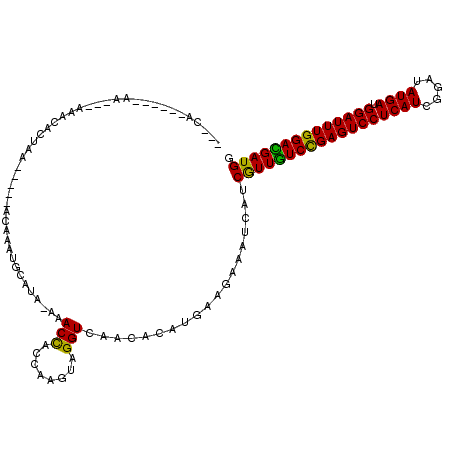
| Location | 17,775,030 – 17,775,133 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.04 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -22.86 |
| Energy contribution | -21.78 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17775030 103 - 22224390 ---CA------AA---ACACACUAA-----ACAAAUGCAUACAAACCACCAAGUAGGUCAACACAUGAAGAAAUCAUCAUUGUCUGAGUCCUCAUCGGAUAUGAUGGAUUUGGACGAUGG ---..------..---.........-----.................(((.....)))......((((.....))))(((((((..(((((((((.....)))).)))))..))))))). ( -22.90) >DroVir_CAF1 49328 113 - 1 AAACAACGUUGAACAUCAACACUAA-----ACAAAUGCAUA-AAACCA-CAAGUAGGUCAACACUUGAAGAAAUCAUCGUUGUCCGAGUCCUCAUCGGAUAUGAUGGAUUUGGACGAUGG .......((((.....)))).....-----...........-......-(((((.(.....))))))..........((((((((((((((((((.....)))).)))))))))))))). ( -31.20) >DroPse_CAF1 44042 103 - 1 ---GA------CA---ACACACUAA-----ACAAUUGACUACAAACUACCAAGUAGGUCAACACAUGAAGAAAUCGUCGUUGUCCGAGUCCUCAUCGGAUAUGAUGGAUUUGGAUGAUGG ---((------(.---.........-----....((((((((..........))).))))).....((.....)))))((..(((((((((((((.....)))).)))))))))..)).. ( -26.00) >DroWil_CAF1 48605 109 - 1 ---CA------AC-UGCAACACUAAAACCAACAAUAACAUA-ACACUACCAAGUAGGUCAACACAUGAAGAAAUCAUCGUUAUCUGAGUCCUCAUCGGAUAUGAUGGAUUUGGAUGAUGG ---..------..-.......................(((.-.....(((.....)))......)))..........(((((((..(((((((((.....)))).)))))..))))))). ( -21.60) >DroMoj_CAF1 42307 110 - 1 AAACAACGUUCAA---CAACACUAA-----ACAAAUGCAUA-AAACUA-CAAGUAGGUCAACACUUGAAGAAAUCAUCGUUGUCCGAGUCCUCAUCGGAUAUGAUGGAUUUGGAUGAUGG ...((.(((((((---.........-----...........-...((.-(((((.(.....)))))).))...(((((((.((((((.......)))))))))))))..))))))).)). ( -26.80) >DroAna_CAF1 40016 104 - 1 ---CA------AA-C-ACACACUAA-----ACCAAUGCAUACACACCACCAAGUAGGUCAACACAUGAAGAAAUCAUCAUUGUCCGAGUCCUCAUCGGAUAUGAUGGAUUUGGACGAUGG ---..------..-.-.........-----.................(((.....)))......((((.....))))((((((((((((((((((.....)))).)))))))))))))). ( -25.40) >consensus ___CA______AA___AAACACUAA_____ACAAAUGCAUA_AAACCACCAAGUAGGUCAACACAUGAAGAAAUCAUCGUUGUCCGAGUCCUCAUCGGAUAUGAUGGAUUUGGACGAUGG ............................................(((........)))...................((((((((((((((((((.....)))).)))))))))))))). (-22.86 = -21.78 + -1.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:49 2006