
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,767,873 – 17,768,004 |
| Length | 131 |
| Max. P | 0.988421 |

| Location | 17,767,873 – 17,767,977 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -22.50 |
| Energy contribution | -22.38 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17767873 104 - 22224390 AGAUCUAAGUCCAGAUCCUUCCCUACGAAAAGGGGGGCUUGGCACCUCCCCUUUUCUUGUCGAACACGCAGACAGACAACCCUACACACACACACCCGAGUCAU .(((((......))))).........(((((((((((........))))))))))).((((.........))))(((......................))).. ( -24.85) >DroSec_CAF1 26794 102 - 1 AGAUCUAAGUCCAGACCCUUCCCUCCGAAAAGGGGGGCUUGGCACCUCCCCUUUUCUUUUCGGACACACAGACAGACCACCCCAC--ACACACACUCGAGUCAU ........((((.((....)).....(((((((((((........))))))))))).....)))).....(((.((.........--........))..))).. ( -23.53) >DroEre_CAF1 25193 101 - 1 AGAUCUAAGUCCAGACCCUUCGCUCUGAAAAGGGGGGCUUGGCACCUCCCCUUUUCUUGUCGAACACGCAGACAGACAACCC---ACACACAUACCCGAGUCAU .....................((((.(((((((((((........)))))))))))(((((.........))))).......---............))))... ( -24.30) >DroYak_CAF1 25532 104 - 1 AGAUCUAAGUCCAGACCCUUCGUUUCGAAGAGGGGGGCUUGGCACCUCCCCUUUUCUUGUCGAACACGCAGACAGACAACCCCAUACACACAUACCCGAGUCAU .............(((...(((....(((((((((((........)))))))))))(((((.........))))).....................)))))).. ( -24.80) >consensus AGAUCUAAGUCCAGACCCUUCCCUCCGAAAAGGGGGGCUUGGCACCUCCCCUUUUCUUGUCGAACACGCAGACAGACAACCCCACACACACACACCCGAGUCAU .............(((.(........(((((((((((........)))))))))))(((((.........)))))......................).))).. (-22.50 = -22.38 + -0.12)

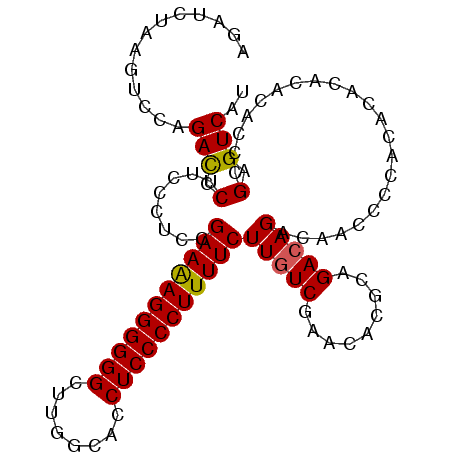
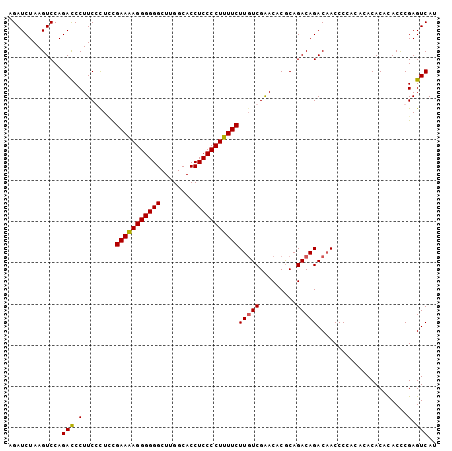
| Location | 17,767,907 – 17,768,004 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 87.16 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -28.84 |
| Energy contribution | -29.40 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17767907 97 + 22224390 UGCGUGUUCGACAAGAAAAGGGGAGGUGCCAAGCCCCCCUUUUCGUAGGGAAGGAUCUGGACUUAGAUCUAGGUAACUAGAGCCAAGGCUUAUAAUU----------- .((.(((((.....(((((((((.(((.....)))))))))))).(((..(.(((((((....)))))))...)..)))))))..).))........----------- ( -31.90) >DroSec_CAF1 26826 97 + 1 UGUGUGUCCGAAAAGAAAAGGGGAGGUGCCAAGCCCCCCUUUUCGGAGGGAAGGGUCUGGACUUAGAUCUAGGUUACUAGAGACAAGGCUUAUAAUU----------- ..(((((((.....(((((((((.(((.....))))))))))))....(((....))))))).....(((((....))))).)))............----------- ( -31.60) >DroEre_CAF1 25224 108 + 1 UGCGUGUUCGACAAGAAAAGGGGAGGUGCCAAGCCCCCCUUUUCAGAGCGAAGGGUCUGGACUUAGAUCUAGGUAACUAGAGCCAAGGCUUAUAAUUAUAAUAUAAGA ......((((....(((((((((.(((.....))))))))))))....))))((.((((((((((....)))))..))))).))....((((((.......)))))). ( -37.30) >DroYak_CAF1 25566 101 + 1 UGCGUGUUCGACAAGAAAAGGGGAGGUGCCAAGCCCCCCUCUUCGAAACGAAGGGUCUGGACUUAGAUCUAGGUAACUAGCGCCAAGGCUUAUGAUUA-------CGA ..((((((((....(((.(((((.(((.....)))))))).)))....)))).((((((((((((....)))))..)))).)))............))-------)). ( -30.60) >consensus UGCGUGUUCGACAAGAAAAGGGGAGGUGCCAAGCCCCCCUUUUCGGAGCGAAGGGUCUGGACUUAGAUCUAGGUAACUAGAGCCAAGGCUUAUAAUU___________ .((...........(((((((((.(((.....))))))))))))........((.((((((((((....)))))..))))).))...))................... (-28.84 = -29.40 + 0.56)

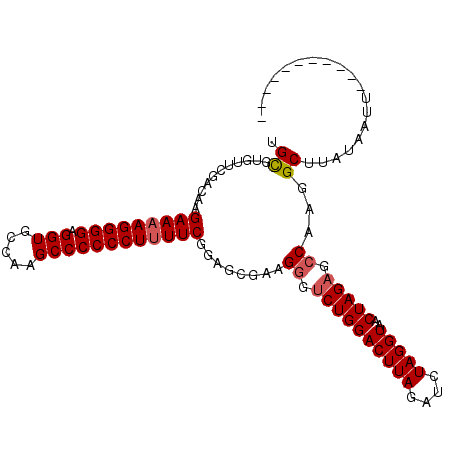
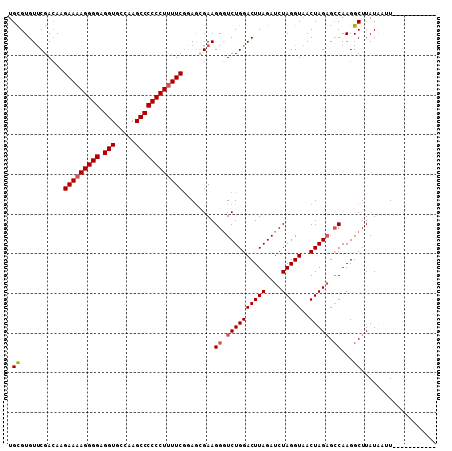
| Location | 17,767,907 – 17,768,004 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.16 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -25.50 |
| Energy contribution | -26.38 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17767907 97 - 22224390 -----------AAUUAUAAGCCUUGGCUCUAGUUACCUAGAUCUAAGUCCAGAUCCUUCCCUACGAAAAGGGGGGCUUGGCACCUCCCCUUUUCUUGUCGAACACGCA -----------........(((((((.(((((....))))).))))).................(((((((((((........)))))))))))...........)). ( -28.30) >DroSec_CAF1 26826 97 - 1 -----------AAUUAUAAGCCUUGUCUCUAGUAACCUAGAUCUAAGUCCAGACCCUUCCCUCCGAAAAGGGGGGCUUGGCACCUCCCCUUUUCUUUUCGGACACACA -----------................(((((....))))).....((((.((....)).....(((((((((((........))))))))))).....))))..... ( -26.00) >DroEre_CAF1 25224 108 - 1 UCUUAUAUUAUAAUUAUAAGCCUUGGCUCUAGUUACCUAGAUCUAAGUCCAGACCCUUCGCUCUGAAAAGGGGGGCUUGGCACCUCCCCUUUUCUUGUCGAACACGCA .((((((.......)))))).(((((.(((((....))))).))))).........(((((...(((((((((((........)))))))))))..).))))...... ( -31.90) >DroYak_CAF1 25566 101 - 1 UCG-------UAAUCAUAAGCCUUGGCGCUAGUUACCUAGAUCUAAGUCCAGACCCUUCGUUUCGAAGAGGGGGGCUUGGCACCUCCCCUUUUCUUGUCGAACACGCA ...-------.........(((((((..((((....))))..))))).........((((....(((((((((((........)))))))))))....))))...)). ( -28.90) >consensus ___________AAUUAUAAGCCUUGGCUCUAGUUACCUAGAUCUAAGUCCAGACCCUUCCCUCCGAAAAGGGGGGCUUGGCACCUCCCCUUUUCUUGUCGAACACGCA ...................(((((((.(((((....))))).))))).........((((....(((((((((((........)))))))))))....))))...)). (-25.50 = -26.38 + 0.88)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:46 2006