
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,756,486 – 17,756,586 |
| Length | 100 |
| Max. P | 0.520767 |

| Location | 17,756,486 – 17,756,586 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -46.23 |
| Consensus MFE | -31.44 |
| Energy contribution | -31.75 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
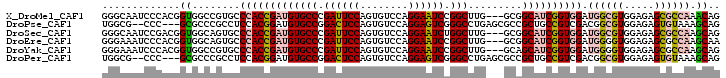
>X_DroMel_CAF1 17756486 100 + 22224390 GGGCAAUCCCACGGUGGCCGUGCCCACCGAUGUGCCCGAUUCCAGUGUCCAGGAAUCCGGCUUG---GCGGCAUCGGUGGAUGGCGUGGAGAGCGCCAAACAG .(((..((((((....(((((..(((((((((((((.((((((........)))))).)))...---...))))))))))))))))))).))..)))...... ( -48.20) >DroPse_CAF1 13791 98 + 1 UGGCG--CCC---GCGCCCGCCUCCACGGAUGUGCCGGACUCCAGUGUCCAGGAGUCGGGCCUGAGCGCCGCUGCCGUCGACGGCGUGGAGAGUGUAAAGCAG .((((--...---.))))(((((((((((.((((((.((((((........)))))).)))....)))))...((((....)))))))))).)))........ ( -47.80) >DroSec_CAF1 15284 100 + 1 GGGCAAUCCGACGGUGGCAGUGCCCACCGAUGUGCCCGAUUCCAGUGUCCAGGAAUCUGGCUUG---GCGGCAUCGGUGGAUGGCGUGGAGAGCGCCAAGCAG (((((...((.((((((......)))))).)))))))((((((........))))))..(((((---(((.((((....)))).)((.....))))))))).. ( -46.20) >DroEre_CAF1 14021 100 + 1 GGGAAAUCCCACGGUGGCAGUGCCCACCGAUGUGCCCGAUUCCAGUGUCCAGGAAUCCGGCUUG---GCGGCAUCGGUGGAUGGGGUGGAGAGCGCCAAGCAA (((....)))......((.....(((((((((((((.((((((........)))))).)))...---...)))))))))).(((.((.....)).))).)).. ( -42.80) >DroYak_CAF1 13985 100 + 1 GGGAAAUCCCACGGUGGCCGUGCCCACCGAUGUGCCCGAUUCCAGUGUCCAGGAAUCCGGCUUG---GCAGCAUCGGUGGAUGGGGUGGAGAGCGCCAAGCAG (((....)))((((...))))(((((((((((((((.((((((........)))))).)))...---...)))))))))).(((.((.....)).))).)).. ( -44.60) >DroPer_CAF1 13590 98 + 1 UGGCG--CCC---GCGCCCGCCUCCACGGAUGUGCCGGACUCCAGUGUCCAGGAGUCGGGCCUGAGCGCCGCUGCCGUCGACGGCGUGGAGAGUGUAAAGCAG .((((--...---.))))(((((((((((.((((((.((((((........)))))).)))....)))))...((((....)))))))))).)))........ ( -47.80) >consensus GGGCAAUCCCACGGUGGCCGUGCCCACCGAUGUGCCCGAUUCCAGUGUCCAGGAAUCCGGCUUG___GCGGCAUCGGUGGAUGGCGUGGAGAGCGCCAAGCAG .............((........(((((((((((((.((((((........)))))).))).........)))))))))).((((((.....)))))).)).. (-31.44 = -31.75 + 0.31)
| Location | 17,756,486 – 17,756,586 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -43.05 |
| Consensus MFE | -26.99 |
| Energy contribution | -26.88 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
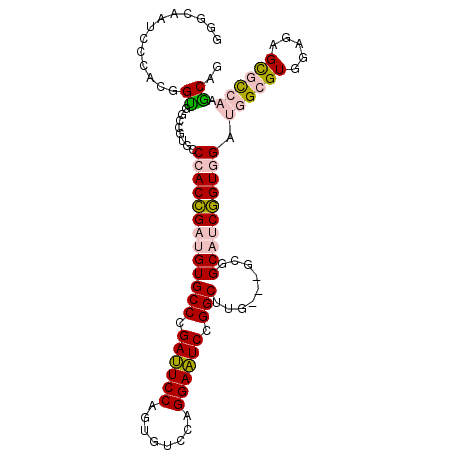
>X_DroMel_CAF1 17756486 100 - 22224390 CUGUUUGGCGCUCUCCACGCCAUCCACCGAUGCCGC---CAAGCCGGAUUCCUGGACACUGGAAUCGGGCACAUCGGUGGGCACGGCCACCGUGGGAUUGCCC ......((((.((.(((((((.((((((((((....---...(((.((((((........)))))).))).))))))))))...)))....)))))).)))). ( -46.10) >DroPse_CAF1 13791 98 - 1 CUGCUUUACACUCUCCACGCCGUCGACGGCAGCGGCGCUCAGGCCCGACUCCUGGACACUGGAGUCCGGCACAUCCGUGGAGGCGGGCGC---GGG--CGCCA ..................((((....))))...(((((((..(((((.((((((((..((((...))))....)))).)))).)))))..---)))--)))). ( -46.70) >DroSec_CAF1 15284 100 - 1 CUGCUUGGCGCUCUCCACGCCAUCCACCGAUGCCGC---CAAGCCAGAUUCCUGGACACUGGAAUCGGGCACAUCGGUGGGCACUGCCACCGUCGGAUUGCCC ..(((((((((.......))((((....))))..))---)))))..((((((........))))))((((((..((((((......))))))..)...))))) ( -41.40) >DroEre_CAF1 14021 100 - 1 UUGCUUGGCGCUCUCCACCCCAUCCACCGAUGCCGC---CAAGCCGGAUUCCUGGACACUGGAAUCGGGCACAUCGGUGGGCACUGCCACCGUGGGAUUUCCC ....((((((..........((((....)))).)))---)))(((.((((((........)))))).)))....((((((......)))))).(((....))) ( -37.40) >DroYak_CAF1 13985 100 - 1 CUGCUUGGCGCUCUCCACCCCAUCCACCGAUGCUGC---CAAGCCGGAUUCCUGGACACUGGAAUCGGGCACAUCGGUGGGCACGGCCACCGUGGGAUUUCCC ..((.....))..(((......((((((((((.(((---(......((((((........)))))).))))))))))))))(((((...))))))))...... ( -40.00) >DroPer_CAF1 13590 98 - 1 CUGCUUUACACUCUCCACGCCGUCGACGGCAGCGGCGCUCAGGCCCGACUCCUGGACACUGGAGUCCGGCACAUCCGUGGAGGCGGGCGC---GGG--CGCCA ..................((((....))))...(((((((..(((((.((((((((..((((...))))....)))).)))).)))))..---)))--)))). ( -46.70) >consensus CUGCUUGGCGCUCUCCACGCCAUCCACCGAUGCCGC___CAAGCCGGAUUCCUGGACACUGGAAUCGGGCACAUCGGUGGGCACGGCCACCGUGGGAUUGCCC ......................((((((((((..(....)..(((.((((((........)))))).))).))))))))))...................... (-26.99 = -26.88 + -0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:40 2006