| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,748,918 – 17,749,012 |
| Length | 94 |
| Max. P | 0.724614 |

| Location | 17,748,918 – 17,749,012 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.07 |
| Mean single sequence MFE | -13.69 |
| Consensus MFE | -9.25 |
| Energy contribution | -9.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
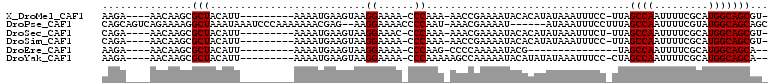
>X_DroMel_CAF1 17748918 94 - 22224390 AAGA----AACAAGCGCUACAUU---------AAAAUGAAGUAAGGAAAA-CCCAAA-AACCGAAAAUACACAUAUAAAUUUCC-UUAGCCAAUUUUCGCAUGGCAGCGU- ....----.....(((((.....---------.........((((((((.-......-.....................)))))-)))((((.........)))))))))- ( -15.69) >DroPse_CAF1 6698 102 - 1 CAGCAGUCAGAAAAGGCUAAAUAAAUCCCAAAAAAACGAG--AAGGAAAACCCCAAU-AAACGAAAAU------AUAAAUUUCCUUUAGCCAAUUUUCGUAUGGCAGCAGC ..((.((((((((((((((((...................--..((......))...-....((((..------.....)))).)))))))..)))))...)))).))... ( -16.90) >DroSec_CAF1 6533 94 - 1 CAGA----AACAAGCGCUACAUU---------AAAAUGAAGUAAGGAAAC-CCCAAA-AAACGAAAAUACACAUAUAAAUUUCU-UUAGCCAAUUUUCGCAUGGCAGCGU- ....----.....(((((.....---------.........((((((((.-......-.....................)))))-)))((((.........)))))))))- ( -12.79) >DroSim_CAF1 6491 94 - 1 CAGA----AACAAGCGCUACAUU---------AAAAUGAAGUAAGGAAAA-CCCAAA-AACCGAAAAUACACAUAUAAAUUUCC-UUAGCCAAUUUUCGCAUGGCAGCGU- ....----.....(((((.....---------.........((((((((.-......-.....................)))))-)))((((.........)))))))))- ( -15.69) >DroEre_CAF1 6727 79 - 1 AAGA----AACAAGCGCUACAUU---------AAAAUGAAGUAAGGAAAA-CCCAAG-CCCCAAAAAUACG---------------UAGCCAAUUUUCGCAUGGCAGCA-- ....----.....(((((.(((.---------...))).)))..((....-.))..)-)...........(---------------(.((((.........)))).)).-- ( -10.80) >DroYak_CAF1 6537 94 - 1 AAGA----AACAAGCGCUACAUU---------AAAAUGAAGUAAGGAAAA-CCCAAAAAGCCAAAAAUACAUAUAUAAAUUUCC-CUAGCCAAUUUUCGCAUGGCAGCA-- ....----.....(((((.(((.---------...)))......((....-.))....))).......................-...((((.........)))).)).-- ( -10.30) >consensus AAGA____AACAAGCGCUACAUU_________AAAAUGAAGUAAGGAAAA_CCCAAA_AACCGAAAAUACACAUAUAAAUUUCC_UUAGCCAAUUUUCGCAUGGCAGCAU_ ...............(((..........................((......))..................................((((.........)))))))... ( -9.25 = -9.25 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:38 2006