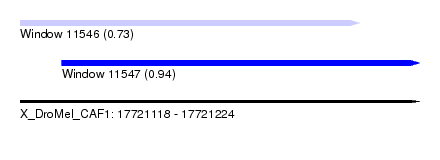
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,721,118 – 17,721,224 |
| Length | 106 |
| Max. P | 0.942175 |

| Location | 17,721,118 – 17,721,208 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 83.63 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -21.40 |
| Energy contribution | -22.52 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17721118 90 + 22224390 GAGCUGGGUUAGCCACUCGCGAUGAUAUCGAUGUAUCGGUGUAUCGGGCGGAACGGUGGUGGUAGACAGCCACAAUCGGGGCAUCGGUUG .((((((....(((.(.(.((((.(((((((....))))))))))).).)...((((.(((((.....))))).)))).))).)))))). ( -32.30) >DroSec_CAF1 25478 90 + 1 GAGCUGGGUUAGCCACUCGCGAUGAUAUCGAUGUAUCGGUGUAUCGGGCGGAACGGUGGUGGUAGCUAGCCACUAUCGGGGCAUCGGUUG .((((((....(((.(.(.((((.(((((((....))))))))))).).)...((((((((((.....)))))))))).))).)))))). ( -36.50) >DroSim_CAF1 27553 90 + 1 GAGCUGGGUUAGCCACUCGCGAUGAUAUCGAUGUAUCGGUGUAUCGGGCGGAACGGUGGUGGUAGAUAGCCACUAUCGGGGCAUCGGUUG .((((((....(((.(.(.((((.(((((((....))))))))))).).)...((((((((((.....)))))))))).))).)))))). ( -36.50) >DroEre_CAF1 26704 82 + 1 GAGCUGGGUUAGCCACUCGCGAGGAUAUCGAUGUA--------UCGGGUGAAAUGGUGGUGGUUGCCUGGCUCUAUCGGGGUUUCGUGUU ((((..(((.((((((.(((.(.....((((....--------))))......).))))))))))))..))))................. ( -30.10) >DroYak_CAF1 26703 82 + 1 GAGCUGGGUUAGCCACUAGCGAUGAUAUCGAUGUA--------UCGGGUGGAGUGGUGGUGGUUGCUAGGCACUAUCGGCGUUUUGGUUU ((((..((...((((((.((..((((((....)))--------))).))..)))(((((((.(.....).))))))))))..))..)))) ( -24.60) >consensus GAGCUGGGUUAGCCACUCGCGAUGAUAUCGAUGUAUCGGUGUAUCGGGCGGAACGGUGGUGGUAGCUAGCCACUAUCGGGGCAUCGGUUG (((.(((.....)))))).(((((...((((((........))))))......((((((((((.....))))))))))...))))).... (-21.40 = -22.52 + 1.12)


| Location | 17,721,129 – 17,721,224 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -36.06 |
| Consensus MFE | -22.73 |
| Energy contribution | -24.69 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17721129 95 + 22224390 GCCACUCGCGAUGAUAUCGAUGUAUCGGUGUAUCGGGCGGAACGGUGGUGGUAGACAGCCACAAUCGGGGCAUCGGUUGCAAUAGUAGCCGAUGG (((.(.(.((((.(((((((....))))))))))).).)...((((.(((((.....))))).)))).)))(((((((((....))))))))).. ( -40.40) >DroSec_CAF1 25489 95 + 1 GCCACUCGCGAUGAUAUCGAUGUAUCGGUGUAUCGGGCGGAACGGUGGUGGUAGCUAGCCACUAUCGGGGCAUCGGUUGCGAUAGUAGCCGAUAU (((.(.(.((((.(((((((....))))))))))).).)...((((((((((.....)))))))))).)))(((((((((....))))))))).. ( -45.30) >DroSim_CAF1 27564 95 + 1 GCCACUCGCGAUGAUAUCGAUGUAUCGGUGUAUCGGGCGGAACGGUGGUGGUAGAUAGCCACUAUCGGGGCAUCGGUUGCGAUAGUAGACGAUGG (((.(.(.((((.(((((((....))))))))))).).)...((((((((((.....)))))))))).)))((((.((((....)))).)))).. ( -38.40) >DroEre_CAF1 26715 87 + 1 GCCACUCGCGAGGAUAUCGAUGUA--------UCGGGUGAAAUGGUGGUGGUUGCCUGGCUCUAUCGGGGUUUCGUGUUCGAUAGUAGCCGAUGG (((((.(((.(.....((((....--------))))......).))))))))....(((((((((((((........)))))))).))))).... ( -28.00) >DroYak_CAF1 26714 87 + 1 GCCACUAGCGAUGAUAUCGAUGUA--------UCGGGUGGAGUGGUGGUGGUUGCUAGGCACUAUCGGCGUUUUGGUUUCGAUAGUAGCCGAUGA (((((((.((......((((....--------))))......)).))))))).....(((((((((((..(....)..)))))))).)))..... ( -28.20) >consensus GCCACUCGCGAUGAUAUCGAUGUAUCGGUGUAUCGGGCGGAACGGUGGUGGUAGCUAGCCACUAUCGGGGCAUCGGUUGCGAUAGUAGCCGAUGG (((..((((.......((((((........))))))))))..((((((((((.....)))))))))).))).((((((((....))))))))... (-22.73 = -24.69 + 1.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:25 2006