
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,719,480 – 17,719,599 |
| Length | 119 |
| Max. P | 0.999813 |

| Location | 17,719,480 – 17,719,599 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -31.42 |
| Energy contribution | -30.70 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17719480 119 + 22224390 AGUGUGGAAAAGCCAUCUUGUCCAGCGGAAGAACGAAUGAA-AAUCACAGCUGCCAAUGCGAGGCUGCCAGCUGUUUGGAUUUUUCCUCCUUUUCCCUUAUGUAUUUGUUUUUUGGAAAA ....((((.(((....))).)))).(.((((((((((((..-....(((((((.((..((...)))).)))))))..(((....)))...............)))))))))))).).... ( -28.00) >DroSec_CAF1 23815 120 + 1 AGUGUGGAAAACCCAUCUUGUCCAGCGGAAGAACGAAGGAAAAAUCACAGCUGCCAAUGCGAUGCUGCCAGCUGUUUGGAUUUUUCCUCCUUUUCCCUUUUGUAUUCGUUUUUCGGAAAA ...((((.....)))).........(.((((((((((((((((((((((((((.((..((...)))).)))))))...)))))))))).................))))))))).).... ( -33.12) >DroSim_CAF1 25904 120 + 1 AGUGUGGAAAACCCAUCUUGUCCAGCGGAAGAACGAAGGAAAAAUCACAGCUGCCAAUGCGAUGCUGCCAGCUGUUUGGAUUUUUCCUCCUUUUCCCUUUUGUAUUCGUUUUUCGGAAAA ...((((.....)))).........(.((((((((((((((((((((((((((.((..((...)))).)))))))...)))))))))).................))))))))).).... ( -33.12) >DroEre_CAF1 24968 119 + 1 AGGGUGGAAAACCCACUUU-UUCAACGGAAAAACGAAGGAAAAAUCACAGCUGCCAAUGCGAUACUGCCAGCUGUUUGGAUUUUUCCUCAUUUUCCCUUUUGUAUUUGUUUUUCGGAAAA .((((.....)))).....-.....(.((((((((((((((((((((((((((.((.........)).)))))))...))))))))).((..........))..)))))))))).).... ( -35.00) >DroYak_CAF1 25004 120 + 1 AGAGUGGAAAACCCAUGUUGUCCAGCGGAAAAGCGAAGGGAAAAUCACAGCUGCCAAUGCGAUGCUGCCAGCUGUUUGGAUUUUUCCUCCUUUUCCCUUUUGUAUUCGUUUUUCGGAAAA ....((((.(((....))).))))..(((((((.((.((((((((((((((((.((..((...)))).)))))))...))))))))))))))))))........((((.....))))... ( -37.10) >consensus AGUGUGGAAAACCCAUCUUGUCCAGCGGAAGAACGAAGGAAAAAUCACAGCUGCCAAUGCGAUGCUGCCAGCUGUUUGGAUUUUUCCUCCUUUUCCCUUUUGUAUUCGUUUUUCGGAAAA ...((((.....)))).........(.((((((((((((((((((((((((((.((.........)).)))))))...)))))))))).................))))))))).).... (-31.42 = -30.70 + -0.72)


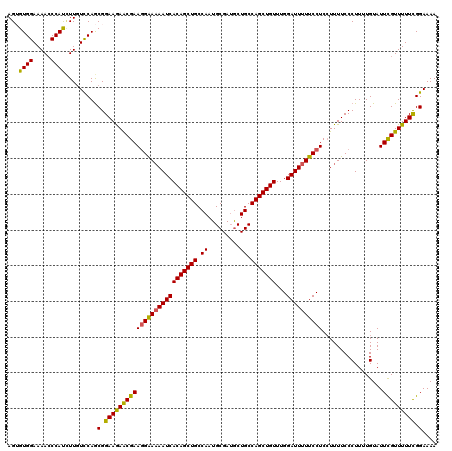
| Location | 17,719,480 – 17,719,599 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -37.26 |
| Consensus MFE | -32.38 |
| Energy contribution | -33.14 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17719480 119 - 22224390 UUUUCCAAAAAACAAAUACAUAAGGGAAAAGGAGGAAAAAUCCAAACAGCUGGCAGCCUCGCAUUGGCAGCUGUGAUU-UUCAUUCGUUCUUCCGCUGGACAAGAUGGCUUUUCCACACU ........................((((((((((..((((((...(((((((.(((.......))).)))))))))))-))..)))((((.......)))).......)))))))..... ( -31.30) >DroSec_CAF1 23815 120 - 1 UUUUCCGAAAAACGAAUACAAAAGGGAAAAGGAGGAAAAAUCCAAACAGCUGGCAGCAUCGCAUUGGCAGCUGUGAUUUUUCCUUCGUUCUUCCGCUGGACAAGAUGGGUUUUCCACACU ........................((((((((((((((((((...(((((((.(((.......))).)))))))))))))))))))((((.......))))........))))))..... ( -38.00) >DroSim_CAF1 25904 120 - 1 UUUUCCGAAAAACGAAUACAAAAGGGAAAAGGAGGAAAAAUCCAAACAGCUGGCAGCAUCGCAUUGGCAGCUGUGAUUUUUCCUUCGUUCUUCCGCUGGACAAGAUGGGUUUUCCACACU ........................((((((((((((((((((...(((((((.(((.......))).)))))))))))))))))))((((.......))))........))))))..... ( -38.00) >DroEre_CAF1 24968 119 - 1 UUUUCCGAAAAACAAAUACAAAAGGGAAAAUGAGGAAAAAUCCAAACAGCUGGCAGUAUCGCAUUGGCAGCUGUGAUUUUUCCUUCGUUUUUCCGUUGAA-AAAGUGGGUUUUCCACCCU ..................(((..(((((((((((((((((((...(((((((.((((.....)))).)))))))))))))))).)))))))))).)))..-.....((((.....)))). ( -42.90) >DroYak_CAF1 25004 120 - 1 UUUUCCGAAAAACGAAUACAAAAGGGAAAAGGAGGAAAAAUCCAAACAGCUGGCAGCAUCGCAUUGGCAGCUGUGAUUUUCCCUUCGCUUUUCCGCUGGACAACAUGGGUUUUCCACUCU .......................((((((.(((((.((((((...(((((((.(((.......))).))))))))))))).)))))..))))))..((((.(((....))).)))).... ( -36.10) >consensus UUUUCCGAAAAACGAAUACAAAAGGGAAAAGGAGGAAAAAUCCAAACAGCUGGCAGCAUCGCAUUGGCAGCUGUGAUUUUUCCUUCGUUCUUCCGCUGGACAAGAUGGGUUUUCCACACU ........................((((((((((((((((((...(((((((.(((.......))).)))))))))))))))))))......(((((.....)).))).))))))..... (-32.38 = -33.14 + 0.76)


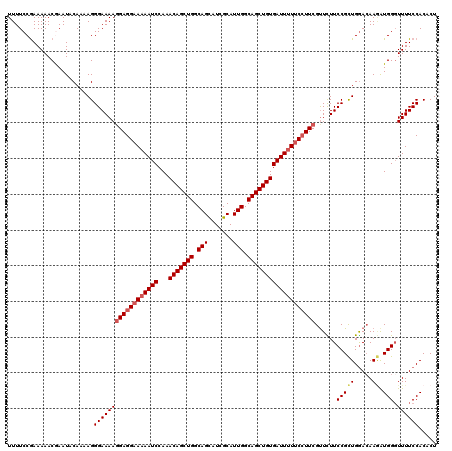
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:21 2006