
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,717,327 – 17,717,478 |
| Length | 151 |
| Max. P | 0.799326 |

| Location | 17,717,327 – 17,717,443 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 88.14 |
| Mean single sequence MFE | -35.36 |
| Consensus MFE | -25.36 |
| Energy contribution | -25.56 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17717327 116 - 22224390 AAUUGAGCUGAGUGGUUGC-CCAUACUUUUCCACCAGCGCUUUUCCAGCUUUCCCAUUUCCCAUUGUCCUGCAGGAUAAAUGGCUGCCAUAGCGAUGGGUAAUCAGCACCUCAGUUG (((((((.((..(((((((-((((..........(((((((.....)))............((((((((....)))).))))))))........))))))))))).)).))))))). ( -36.87) >DroSec_CAF1 21718 116 - 1 AAUUGAGCUGAGUGGUUGC-CCAUACUUUUCCGCCAGCGCUUUUCCAGCUUUCCCACUUCCCAUUGUCCUGCAGGAUAAAUGGCUGCCAUGGCGAUGGGUAAUCAGCACCUCAGUUA (((((((.((..(((((((-((((.......((((((((((.....)))...........(((((((((....)))).)))))..))..)))))))))))))))).)).))))))). ( -41.61) >DroSim_CAF1 23822 116 - 1 AAUUGAGCUGAGUGGUUGC-CCAUACUUUUCCGCCAGCGCUUUUCCAGCUUUCCCACUUCCCAUUGUCCUGCAGGAUAAAUGGCUGCCAUGGCGAUGGGUAAUCAGCACCUCAGUUA (((((((.((..(((((((-((((.......((((((((((.....)))...........(((((((((....)))).)))))..))..)))))))))))))))).)).))))))). ( -41.61) >DroEre_CAF1 22855 116 - 1 AAUUGAGCAGAGUGGUUGC-UCAUACUUCUCCGCCAGCGCUUUUCCUGCAUUCCCAUAUCCCAUUUUCCUGCAGGAUUAAUGGCUGCCAUAGCGAUGUGUAAUUAGCACCUCAGUUG (((((((..(..(((((((-.(((........((((.......(((((((...................)))))))....))))(((....)))))).))))))).)..))))))). ( -28.91) >DroYak_CAF1 22942 116 - 1 AAUUGAGCAAAGUGGUUGCCUCAUACUUUUACGCCAGCGUUAUUCCCAU-UUUCCAUUUUCCAUUUUCCUGGAGGAUUAAUGGCUGUCAGAACGAUGGGUAAUUAGCAGCUCUGUUG (((.((((((((((.........)))))).......((...((((((((-((((......(((((.(((....)))..)))))......))).)))))).)))..)).)))).))). ( -27.80) >consensus AAUUGAGCUGAGUGGUUGC_CCAUACUUUUCCGCCAGCGCUUUUCCAGCUUUCCCAUUUCCCAUUGUCCUGCAGGAUAAAUGGCUGCCAUAGCGAUGGGUAAUCAGCACCUCAGUUG .....((((((((((((((.((((........((((..((.......))..............((((((....)))))).))))(((....))))))))))))))....))))))). (-25.36 = -25.56 + 0.20)



| Location | 17,717,367 – 17,717,478 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -18.98 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17717367 111 - 22224390 ----CGGAAAA-GCAAUUGUUUGCCCUUUAACGAUUUUUUAAUUGAGCUGAGUGGUUGC-CCAUACUUUUCCACCAGCGCUUUUCCAGCUUUCCCAUUUCCCAUUGUCCUGCAGGAU ----.((((((-(((((((((........)))))))..........((((.((((....-..........))))))))))))))))...................((((....)))) ( -30.34) >DroSec_CAF1 21758 111 - 1 ----CGGAAAA-GCACUUGUUUAUCCUUUAACGAUUUUUUAAUUGAGCUGAGUGGUUGC-CCAUACUUUUCCGCCAGCGCUUUUCCAGCUUUCCCACUUCCCAUUGUCCUGCAGGAU ----.((((((-((.................(((((....))))).((((.((((....-..........))))))))))))))))...................((((....)))) ( -28.54) >DroSim_CAF1 23862 111 - 1 ----CGGAAAA-GCACUUGUUUAUCCUUUAACGAUUUUUUAAUUGAGCUGAGUGGUUGC-CCAUACUUUUCCGCCAGCGCUUUUCCAGCUUUCCCACUUCCCAUUGUCCUGCAGGAU ----.((((((-((.................(((((....))))).((((.((((....-..........))))))))))))))))...................((((....)))) ( -28.54) >DroEre_CAF1 22895 115 - 1 UCUGCGGAAAA-GCACUUGCUUUUCCUUUAACGAUUUUUUAAUUGAGCAGAGUGGUUGC-UCAUACUUCUCCGCCAGCGCUUUUCCUGCAUUCCCAUAUCCCAUUUUCCUGCAGGAU (((((((((((-((....)))))))).....(((((....))))).)))))(((.(.((-............)).).)))...(((((((...................))))))). ( -32.81) >DroYak_CAF1 22982 113 - 1 ---GCGGAAAGGGCACUUGUUUGCCCUUUAACGAUUUUUUAAUUGAGCAAAGUGGUUGCCUCAUACUUUUACGCCAGCGUUAUUCCCAU-UUUCCAUUUUCCAUUUUCCUGGAGGAU ---..((((((((((......)))))..(((((......(((.(((((((.....))).)))).....)))......))))).......-)))))((((((((......)))))))) ( -26.60) >consensus ____CGGAAAA_GCACUUGUUUAUCCUUUAACGAUUUUUUAAUUGAGCUGAGUGGUUGC_CCAUACUUUUCCGCCAGCGCUUUUCCAGCUUUCCCAUUUCCCAUUGUCCUGCAGGAU .....((((((.((.................(((((....))))).((((.((((...............))))))))))))))))....................(((....))). (-18.98 = -19.50 + 0.52)

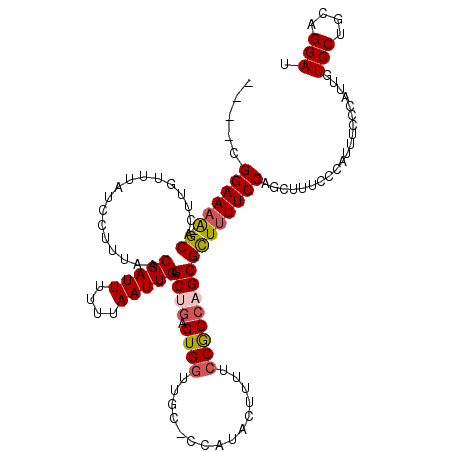
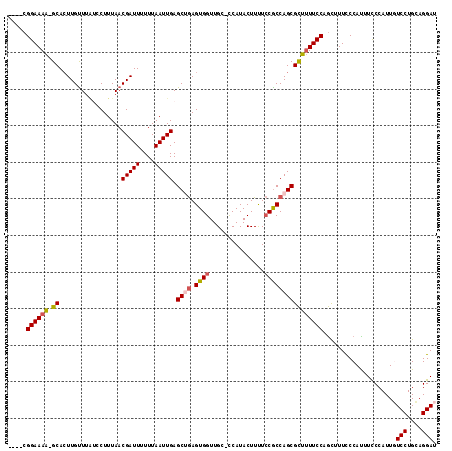
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:18 2006