
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,704,016 – 17,704,127 |
| Length | 111 |
| Max. P | 0.549057 |

| Location | 17,704,016 – 17,704,127 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.16 |
| Mean single sequence MFE | -30.25 |
| Consensus MFE | -19.69 |
| Energy contribution | -19.13 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
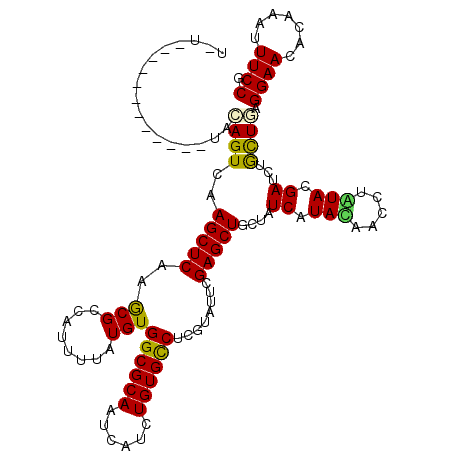
>X_DroMel_CAF1 17704016 111 - 22224390 U-UGAU--------CCGCAGUUAAGCUCAAGCGGCACUUCAUGUGGCGCAACCACCUGUGCCUGGUGUUCGAGCUGCUCUCAUACAACCUGUACGACCUGCUGCGGAACACCAACUUCCG .-((.(--------(((((((..(((((..(((.(((.....))).)))...((((.......))))...)))))....((.(((.....))).))...)))))))).)).......... ( -38.70) >DroVir_CAF1 9925 111 - 1 G-UCAU--------UUGCAGUCAAGCUCAAGCGCCAUUUUAUGUGGCGCAAUCAUCUGUGCCUCGUCUUCGAGCUGCUCUCGUAUAACCUGUACGAUCUGCUUAGGAACACAAAUUUCCG .-....--------..((((...(((((..(((((((.....)))))))...............(....))))))....((((((.....)))))).))))...((((.......)))). ( -31.40) >DroGri_CAF1 11354 105 - 1 ---------------UAUAGUCAAGCUCAAGCGUCAUUUUAUGUGGCGCAAUCAUCUGUGCCUCGUAUUCGAGCUGCUAUCGUAUAACCUAUACGAUCUGCUAAGGAACACAAAUUUCCG ---------------........(((((..((((......))))((((((......))))))........)))))((.((((((((...))))))))..))...((((.......)))). ( -28.20) >DroWil_CAF1 11253 120 - 1 UAUGAUGUCGUCUCUUACAGUCAAGCUCAAACGGCAUUUUAUGUGGCGCAAUCAUUUGUGUCUGGUAUUCGAGCUGCUAUCAUACAACCUAUACGAUCUGCUAAGGAAUACUAAUUUCCG ...((((((((..(((......))).....))))))))......(((((((....)))))))((((((((.(((....(((.............)))..)))...))))))))....... ( -23.82) >DroMoj_CAF1 9054 106 - 1 --------------CUACAGUCAAGCUCAAGCGUCAUUUUAUGUGGCGCAAUCAUCUGUGCCUCGUCUUCGAGCUGCUAUCGUAUAAUCUAUACGAUCUAUUGAGGAACACGAAUUUCCG --------------...((((..((((((((.........(((.((((((......)))))).)))))).)))))...((((((((...))))))))..)))).((((.......)))). ( -26.50) >DroAna_CAF1 10657 107 - 1 U-U------------AACAGUCAAGCUCAAGCGCCAUUUUAUGUGGCGCAAUCACCUGUGCCUGGUGUUCGAGCUGCUCUCAUACAACCUGUACGAUCUGCUGCGGAACACAAACUUCCG .-.------------..((((..(((((..(((((((.....)))))))...((((.......))))...)))))....((.(((.....))).))...))))(((((.......))))) ( -32.90) >consensus U_U____________UACAGUCAAGCUCAAGCGCCAUUUUAUGUGGCGCAAUCAUCUGUGCCUCGUAUUCGAGCUGCUAUCAUACAACCUAUACGAUCUGCUGAGGAACACAAAUUUCCG .................((((..(((((..(((........)))((((((......))))))........)))))....((.(((.....))).))...)))).((((.......)))). (-19.69 = -19.13 + -0.55)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:15 2006