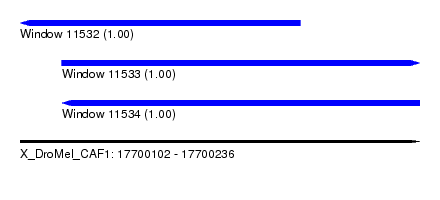
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,700,102 – 17,700,236 |
| Length | 134 |
| Max. P | 0.999937 |

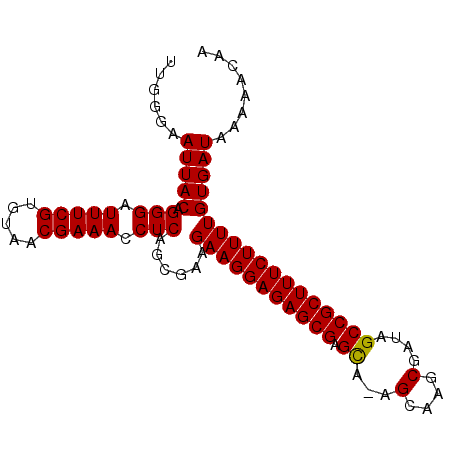
| Location | 17,700,102 – 17,700,196 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -26.50 |
| Energy contribution | -26.34 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.23 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.01 |
| SVM RNA-class probability | 0.999756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17700102 94 - 22224390 UUGGGAAUUACAGGGAUUUCGUGUAACGAAACCUCAGCGAAGAAGGAGAGCGAGCAAAGCAAGCGAUAGCCGCUUUCUUUUGUGAUAAAAACAA ......(((((.(((.(((((.....))))).)))......(((((((((((.((...(....)....))))))))))))))))))........ ( -26.20) >DroSec_CAF1 4263 93 - 1 UUGGGAAUUACAGGGAUUUCGUGUAACGAAACCUCAGCGAAGAAGGAGAGCGAGCA-AGCAAGCGAUAGCCGCUUUCUUUUGUGAUAAAAACAA ......(((((.(((.(((((.....))))).)))......(((((((((((.((.-..(....)...))))))))))))))))))........ ( -26.00) >DroSim_CAF1 5327 93 - 1 UUGGGAAUUACAGGGAUUUCGUGUAACGAAACCUCAGCGAAGAAGGAGAGCGAGCA-AGCAAGCGAUAGCCGCUUUCUUUUGUGAUAAAAACAA ......(((((.(((.(((((.....))))).)))......(((((((((((.((.-..(....)...))))))))))))))))))........ ( -26.00) >DroEre_CAF1 5137 93 - 1 UUGGGGAUUACAGGGAUUUCGUGUAACGAAACCUCAGCGAAGAAGGAGAGCGAGUG-AGCAAGCGGUAGCCGCUUUCUUUUGUGAUAAAAACAA .((...(((((.(((.(((((.....))))).)))......(((((((((((.((.-.((.....)).)))))))))))))))))).....)). ( -26.20) >DroYak_CAF1 5156 93 - 1 UUGGGAAUUACAGGGAUUUCGUGUGACGAAACCUCAGCGAAGAAGGAGAGCGAGCG-AGCAAGCGAUAGCCGCUUUCUUUUGUGAUAAAAACAA ......(((((.(((.(((((.....))))).)))......(((((((((((.((.-..(....)...))))))))))))))))))........ ( -27.10) >consensus UUGGGAAUUACAGGGAUUUCGUGUAACGAAACCUCAGCGAAGAAGGAGAGCGAGCA_AGCAAGCGAUAGCCGCUUUCUUUUGUGAUAAAAACAA ......(((((.(((.(((((.....))))).)))......(((((((((((.((...(....)....))))))))))))))))))........ (-26.50 = -26.34 + -0.16)


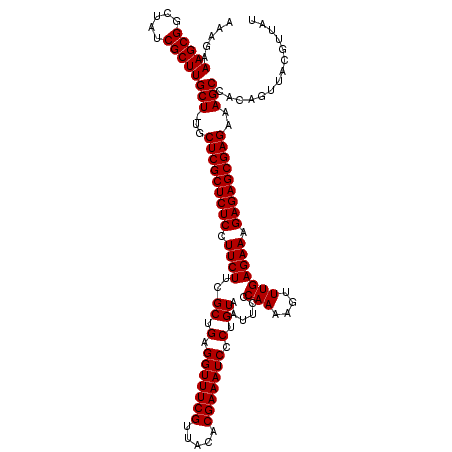
| Location | 17,700,116 – 17,700,236 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.82 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -36.18 |
| Energy contribution | -36.18 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.26 |
| SVM RNA-class probability | 0.999854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17700116 120 + 22224390 AAAGAAAGCGGCUAUCGCUUGCUUUGCUCGCUCUCCUUCUUCGCUGAGGUUUCGUUACACGAAAUCCCUGUAAUUCCCAAAAGUUUGAGAAAGAGAGCGAGAAAGCCACAGUUACGUUAU .....(((((.....)))))(((((.(((((((((.(((((.(((..((....((((((.(.....).))))))..))...)))..))))).)))))))))))))).............. ( -37.80) >DroSec_CAF1 4277 119 + 1 AAAGAAAGCGGCUAUCGCUUGCU-UGCUCGCUCUCCUUCUUCGCUGAGGUUUCGUUACACGAAAUCCCUGUAAUUCCCAAAAGUUUGAGAAAGAGAGCGAGAAAGCCACAGUUACGUUAU .....(((((.....)))))(((-(.(((((((((.(((((.(((..((....((((((.(.....).))))))..))...)))..))))).))))))))).)))).............. ( -37.20) >DroSim_CAF1 5341 119 + 1 AAAGAAAGCGGCUAUCGCUUGCU-UGCUCGCUCUCCUUCUUCGCUGAGGUUUCGUUACACGAAAUCCCUGUAAUUCCCAAAAGUUUGAGAAAGAGAGCGAGAAAGCCACAGUUACGUUAU .....(((((.....)))))(((-(.(((((((((.(((((.(((..((....((((((.(.....).))))))..))...)))..))))).))))))))).)))).............. ( -37.20) >DroEre_CAF1 5151 119 + 1 AAAGAAAGCGGCUACCGCUUGCU-CACUCGCUCUCCUUCUUCGCUGAGGUUUCGUUACACGAAAUCCCUGUAAUCCCCAAAAGUUUGAGAAAGAGAGCGAGAAAGCCACAGUUACGUUAU .....((((((...))))))(((-..(((((((((.(((((.(((..((....((((((.(.....).))))))..))...)))..))))).)))))))))..))).............. ( -37.70) >DroYak_CAF1 5170 119 + 1 AAAGAAAGCGGCUAUCGCUUGCU-CGCUCGCUCUCCUUCUUCGCUGAGGUUUCGUCACACGAAAUCCCUGUAAUUCCCAAAAGUUUGAGAAAGAGAGCGAGAAAGCCACAGUUACGUUAU .....(((((.....)))))(((-..(((((((((.((((..((.(.(((((((.....))))))).).))......(((....))))))).)))))))))..))).............. ( -37.10) >consensus AAAGAAAGCGGCUAUCGCUUGCU_UGCUCGCUCUCCUUCUUCGCUGAGGUUUCGUUACACGAAAUCCCUGUAAUUCCCAAAAGUUUGAGAAAGAGAGCGAGAAAGCCACAGUUACGUUAU .....(((((.....)))))(((...(((((((((.((((..((.(.(((((((.....))))))).).))......(((....))))))).)))))))))..))).............. (-36.18 = -36.18 + -0.00)


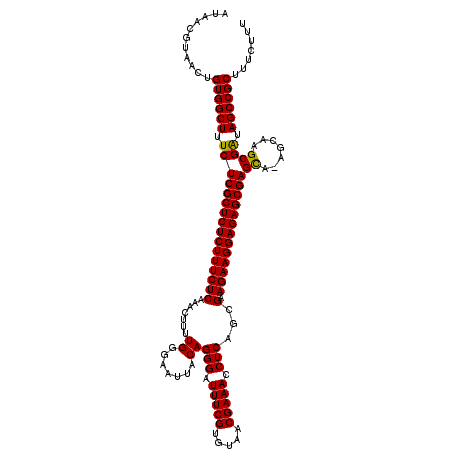
| Location | 17,700,116 – 17,700,236 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.82 |
| Mean single sequence MFE | -42.04 |
| Consensus MFE | -40.62 |
| Energy contribution | -40.30 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.68 |
| SVM RNA-class probability | 0.999937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17700116 120 - 22224390 AUAACGUAACUGUGGCUUUCUCGCUCUCUUUCUCAAACUUUUGGGAAUUACAGGGAUUUCGUGUAACGAAACCUCAGCGAAGAAGGAGAGCGAGCAAAGCAAGCGAUAGCCGCUUUCUUU ..............((((((((((((((((((((.......((.......))(((.(((((.....))))).)))...).)))))))))))))).)))))(((((.....)))))..... ( -42.00) >DroSec_CAF1 4277 119 - 1 AUAACGUAACUGUGGCUUUCUCGCUCUCUUUCUCAAACUUUUGGGAAUUACAGGGAUUUCGUGUAACGAAACCUCAGCGAAGAAGGAGAGCGAGCA-AGCAAGCGAUAGCCGCUUUCUUU ..............((((.(((((((((((((((.......((.......))(((.(((((.....))))).)))...).)))))))))))))).)-)))(((((.....)))))..... ( -41.40) >DroSim_CAF1 5341 119 - 1 AUAACGUAACUGUGGCUUUCUCGCUCUCUUUCUCAAACUUUUGGGAAUUACAGGGAUUUCGUGUAACGAAACCUCAGCGAAGAAGGAGAGCGAGCA-AGCAAGCGAUAGCCGCUUUCUUU ..............((((.(((((((((((((((.......((.......))(((.(((((.....))))).)))...).)))))))))))))).)-)))(((((.....)))))..... ( -41.40) >DroEre_CAF1 5151 119 - 1 AUAACGUAACUGUGGCUUUCUCGCUCUCUUUCUCAAACUUUUGGGGAUUACAGGGAUUUCGUGUAACGAAACCUCAGCGAAGAAGGAGAGCGAGUG-AGCAAGCGGUAGCCGCUUUCUUU ..............((((.(((((((((((((((.......((.......))(((.(((((.....))))).)))...).)))))))))))))).)-)))((((((...))))))..... ( -43.50) >DroYak_CAF1 5170 119 - 1 AUAACGUAACUGUGGCUUUCUCGCUCUCUUUCUCAAACUUUUGGGAAUUACAGGGAUUUCGUGUGACGAAACCUCAGCGAAGAAGGAGAGCGAGCG-AGCAAGCGAUAGCCGCUUUCUUU ...........((((((.((((((((((((((((.......((.......))(((.(((((.....))))).)))...).)))))))))))))((.-.....)))).))))))....... ( -41.90) >consensus AUAACGUAACUGUGGCUUUCUCGCUCUCUUUCUCAAACUUUUGGGAAUUACAGGGAUUUCGUGUAACGAAACCUCAGCGAAGAAGGAGAGCGAGCA_AGCAAGCGAUAGCCGCUUUCUUU ...........((((((.((((((((((((((((.......((.......))(((.(((((.....))))).)))...).)))))))))))))((.......)))).))))))....... (-40.62 = -40.30 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:29:13 2006