| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,649,316 – 17,649,449 |
| Length | 133 |
| Max. P | 0.999081 |

| Location | 17,649,316 – 17,649,421 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.26 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -8.56 |
| Energy contribution | -8.60 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.37 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17649316 105 - 22224390 GAAUCCCAAG--------AUCCCCAGAUCGCAAGAUCUCCAGAUCUCAAGAUCUUCAGAUCUCCAGAUCCCGAAAGGAAAUGCCACGAAAUUGAAAAUGCAAAACGAAAUUGG .....((((.--------.((...(((((....)))))...(((((..(((((....)))))..)))))((....))............................))..)))) ( -26.20) >DroSec_CAF1 42330 96 - 1 G-AUCCCAAG--------AUCCCCAGAU--------CCCCAGAUCGCAAGAUCUCAAGAUCUCCAGAUCCCGAAAGGAAAUGCCACGAAAUUGAAAAUGCAAAACGAAAUUGG .-...(((((--------(((...((((--------(...(((((....)))))...)))))...))))((....))................................)))) ( -24.00) >DroSim_CAF1 10359 80 - 1 G-AUCCCAAG--------AUCCCCAGA----------------UCG--------CAAGAUCUCCAGAUCCCGAAAGGAAAUGCCACGAAAUUGAAAAUGCAAAACGAAAUUGG .-...(((((--------(((...(((----------------((.--------...)))))...))))((....))................................)))) ( -18.70) >DroEre_CAF1 43018 80 - 1 G-AUCCCAAG--------AUCUCCAGC------------------------UCGCGAGAUGCCCAGAUCCCGAAAGGAAGUGCCACAAAAUUGAAAAUGCAAAACGAAAUUGG .-...(((((--------((((...((------------------------((....)).))..)))))((....))................................)))) ( -16.10) >DroYak_CAF1 44086 112 - 1 G-AGCCCAAGAUCCCAAGAUCUCCAGUUCUGCAGAUCUCCAGUUCUGCAGAUCUCCAGAUCUUCAGAUUCCGAAAGGAAAUGCCACGAAAUUGAAAAUGCAAAACGAAAUUGG .-...((((..((..(((((((..((.((((((((........)))))))).))..)))))))....((((....))))..........................))..)))) ( -32.10) >consensus G_AUCCCAAG________AUCCCCAGAU________C_CCAG_UCG__AGAUCUCAAGAUCUCCAGAUCCCGAAAGGAAAUGCCACGAAAUUGAAAAUGCAAAACGAAAUUGG .....((((................................................((((....))))((....))................................)))) ( -8.56 = -8.60 + 0.04)

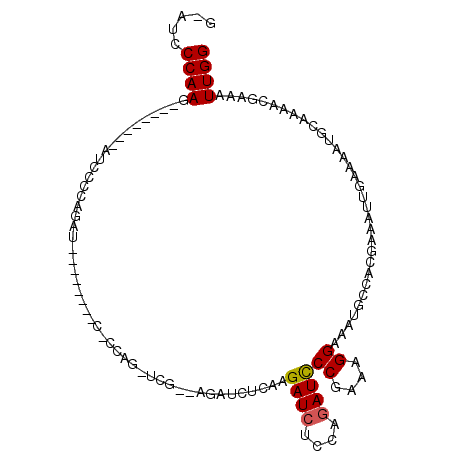
| Location | 17,649,356 – 17,649,449 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -22.90 |
| Energy contribution | -24.01 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.63 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17649356 93 + 22224390 UCGGGAUCUGGAGAUCUGAAGAUCUUGAGAUCUGGAGAUCUUGCGAUCUGGGGAU--------CUUGGGAUUCGAACCUAAGCCGAAAGCUUUAGCCAGCG ...((((((.(((((((..(((((.(((((((....))))))).)))))..))))--------))).))))))....(((((.(....).)))))...... ( -36.90) >DroSec_CAF1 42370 84 + 1 UCGGGAUCUGGAGAUCUUGAGAUCUUGCGAUCUGGGG--------AUCUGGGGAU--------CUUGGGAU-CGAACCUAAGCCGAAAGCUUUAGCCAGCG ..(((((((.((((((((.(((((((........)))--------)))).)))))--------))).))))-)....(((((.(....).))))))).... ( -34.00) >DroYak_CAF1 44126 100 + 1 UCGGAAUCUGAAGAUCUGGAGAUCUGCAGAACUGGAGAUCUGCAGAACUGGAGAUCUUGGGAUCUUGGGCU-CGCACCUAAGCCAAAAGCUUUAGCCAGCG (((..((((.(((((((..((.((((((((.(....).)))))))).))..))))))).))))..)))(((-.((....((((.....))))..)).))). ( -38.70) >consensus UCGGGAUCUGGAGAUCUGGAGAUCUUGAGAUCUGGAGAUCU___GAUCUGGGGAU________CUUGGGAU_CGAACCUAAGCCGAAAGCUUUAGCCAGCG ((.((((((.((((((....)))))).)))))).))...........((((............((((((.......)))))).............)))).. (-22.90 = -24.01 + 1.11)


| Location | 17,649,356 – 17,649,449 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -16.22 |
| Energy contribution | -16.67 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17649356 93 - 22224390 CGCUGGCUAAAGCUUUCGGCUUAGGUUCGAAUCCCAAG--------AUCCCCAGAUCGCAAGAUCUCCAGAUCUCAAGAUCUUCAGAUCUCCAGAUCCCGA .(((......)))..(((((((.((.......)).)))--------......(((((....)))))...(((((..(((((....)))))..))))))))) ( -26.90) >DroSec_CAF1 42370 84 - 1 CGCUGGCUAAAGCUUUCGGCUUAGGUUCG-AUCCCAAG--------AUCCCCAGAU--------CCCCAGAUCGCAAGAUCUCAAGAUCUCCAGAUCCCGA .(((......)))..((((....((....-..))...(--------(((...((((--------(...(((((....)))))...)))))...)))))))) ( -24.40) >DroYak_CAF1 44126 100 - 1 CGCUGGCUAAAGCUUUUGGCUUAGGUGCG-AGCCCAAGAUCCCAAGAUCUCCAGUUCUGCAGAUCUCCAGUUCUGCAGAUCUCCAGAUCUUCAGAUUCCGA .(((.((..((((.....))))....)).-)))....((((..(((((((..((.((((((((........)))))))).))..)))))))..)))).... ( -34.80) >consensus CGCUGGCUAAAGCUUUCGGCUUAGGUUCG_AUCCCAAG________AUCCCCAGAUC___AGAUCUCCAGAUCUCAAGAUCUCCAGAUCUCCAGAUCCCGA ..((((...((((.....)))).((........))...............))))...............(((((..(((((....)))))..))))).... (-16.22 = -16.67 + 0.45)


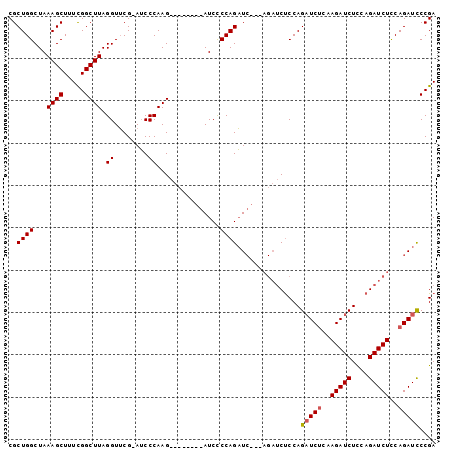
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:58 2006