
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,625,730 – 17,625,851 |
| Length | 121 |
| Max. P | 0.981527 |

| Location | 17,625,730 – 17,625,821 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 88.81 |
| Mean single sequence MFE | -21.85 |
| Consensus MFE | -18.58 |
| Energy contribution | -19.07 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17625730 91 + 22224390 GUUGUUGCUGCCGCCUGUUGUUGAUUAAAAAUUGAUCAAAAAACAUAAAAACACACGAAUUAUGCAUGCGCGUAUUUAAAUGCGUGUGUUG ...............((((.(((((((.....)))))))..))))....((((((((...(((((....)))))........)))))))). ( -19.70) >DroSec_CAF1 21700 87 + 1 ---GUUGCUGCCGCAUGUUGUUGAUUAAAAAUUGAUCAA-AAACAUAAAACCACACGAAUUAUGCAUGCGCGUAUGUAAAUGCGUGUGUUG ---...((....))(((((.(((((((.....)))))))-.))))).....((((((.(((.((((((....))))))))).))))))... ( -23.00) >DroEre_CAF1 24122 74 + 1 ----------------GUUGUUGAUUAAAAAUUGAUCAA-AAACAUAAAAACACACGAAUUAUGCAUGCGCGUAUGUAAAUGCGUGUGUUG ----------------(((.(((((((.....)))))))-.))).....((((((((.(((.((((((....))))))))).)))))))). ( -22.80) >DroYak_CAF1 23151 88 + 1 ---GUUGCUGCCGCCUGUUGUUGAUUAAAAAUUGAUCAAAAAACAUAAAAACACACGAAUUAUGCAUGCGCGUAUGUAAAUGCGUGUGUUG ---............((((.(((((((.....)))))))..))))....((((((((.(((.((((((....))))))))).)))))))). ( -21.90) >consensus ___GUUGCUGCCGCCUGUUGUUGAUUAAAAAUUGAUCAA_AAACAUAAAAACACACGAAUUAUGCAUGCGCGUAUGUAAAUGCGUGUGUUG ................(((..((((((.....))))))...))).....((((((((.(((.((((((....))))))))).)))))))). (-18.58 = -19.07 + 0.50)



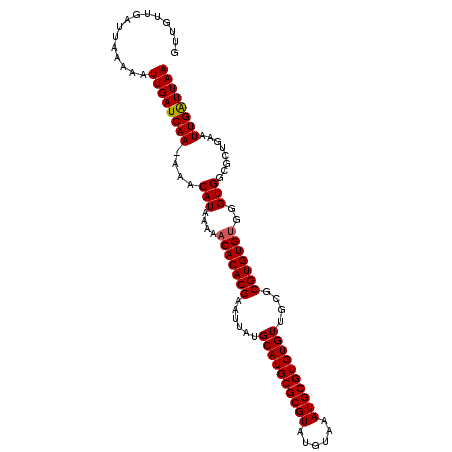
| Location | 17,625,746 – 17,625,851 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 97.93 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -28.74 |
| Energy contribution | -28.80 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17625746 105 + 22224390 GUUGUUGAUUAAAAAUUGAUCAAAAAACAUAAAAACACACGAAUUAUGCAUGCGCGUAUUUAAAUGCGUGUGUUGCGCGUGUGUGGGUGGCGCUGAAUUGAUUAA ....(((((((.....)))))))....(((....(((((((......((((((((((......))))))))))....)))))))..)))................ ( -29.80) >DroSec_CAF1 21713 104 + 1 GUUGUUGAUUAAAAAUUGAUCAA-AAACAUAAAACCACACGAAUUAUGCAUGCGCGUAUGUAAAUGCGUGUGUUGCGCGUGUGUGGGUGGCGCUGAAUUGAUUAA (((.(((((((.....)))))))-.)))......(((((((......((((((((((......))))))))))......)))))))................... ( -31.30) >DroEre_CAF1 24122 104 + 1 GUUGUUGAUUAAAAAUUGAUCAA-AAACAUAAAAACACACGAAUUAUGCAUGCGCGUAUGUAAAUGCGUGUGUUGCGCGUGUGUGGGUGGCGCUGAAUUGGUUAA (((.(((((((.....)))))))-.)))......(((((((......((((((((((......))))))))))....)))))))...((((.(......))))). ( -30.40) >DroYak_CAF1 23164 105 + 1 GUUGUUGAUUAAAAAUUGAUCAAAAAACAUAAAAACACACGAAUUAUGCAUGCGCGUAUGUAAAUGCGUGUGUUGCGCGUGUGUGGGUGGCGCUGAAUUGAUUAA ....(((((((.....)))))))....(((....(((((((......((((((((((......))))))))))....)))))))..)))................ ( -29.80) >consensus GUUGUUGAUUAAAAAUUGAUCAA_AAACAUAAAAACACACGAAUUAUGCAUGCGCGUAUGUAAAUGCGUGUGUUGCGCGUGUGUGGGUGGCGCUGAAUUGAUUAA ...............((((((((....(((....(((((((......((((((((((......))))))))))....)))))))..)))........)))))))) (-28.74 = -28.80 + 0.06)


| Location | 17,625,746 – 17,625,851 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 97.93 |
| Mean single sequence MFE | -18.67 |
| Consensus MFE | -15.73 |
| Energy contribution | -16.22 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17625746 105 - 22224390 UUAAUCAAUUCAGCGCCACCCACACACGCGCAACACACGCAUUUAAAUACGCGCAUGCAUAAUUCGUGUGUUUUUAUGUUUUUUGAUCAAUUUUUAAUCAACAAC ...(((((....((((...........)))).......((((..(((((((((.((.....)).)))))))))..))))...))))).................. ( -20.50) >DroSec_CAF1 21713 104 - 1 UUAAUCAAUUCAGCGCCACCCACACACGCGCAACACACGCAUUUACAUACGCGCAUGCAUAAUUCGUGUGGUUUUAUGUUU-UUGAUCAAUUUUUAAUCAACAAC ...(((((..((((((...........))))..((((((.(((..(((......)))...))).))))))......))...-))))).................. ( -17.80) >DroEre_CAF1 24122 104 - 1 UUAACCAAUUCAGCGCCACCCACACACGCGCAACACACGCAUUUACAUACGCGCAUGCAUAAUUCGUGUGUUUUUAUGUUU-UUGAUCAAUUUUUAAUCAACAAC ............((((...........))))((((((((.(((..(((......)))...))).)))))))).....(((.-(((((.........))))).))) ( -17.80) >DroYak_CAF1 23164 105 - 1 UUAAUCAAUUCAGCGCCACCCACACACGCGCAACACACGCAUUUACAUACGCGCAUGCAUAAUUCGUGUGUUUUUAUGUUUUUUGAUCAAUUUUUAAUCAACAAC ............((((...........))))((((((((.(((..(((......)))...))).))))))))....((((..((((.......))))..)))).. ( -18.60) >consensus UUAAUCAAUUCAGCGCCACCCACACACGCGCAACACACGCAUUUACAUACGCGCAUGCAUAAUUCGUGUGUUUUUAUGUUU_UUGAUCAAUUUUUAAUCAACAAC ............((((...........))))((((((((.(((..(((......)))...))).))))))))....((((..((((.......))))..)))).. (-15.73 = -16.22 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:52 2006