
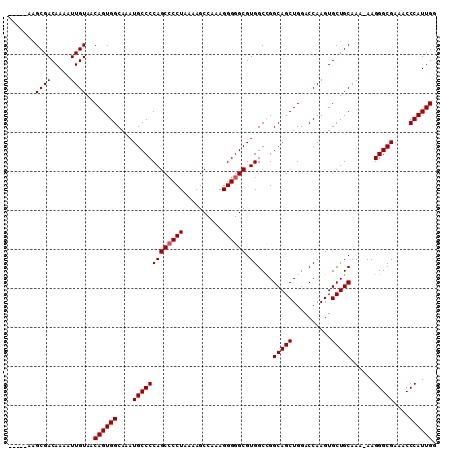
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,593,385 – 17,593,549 |
| Length | 164 |
| Max. P | 0.994706 |

| Location | 17,593,385 – 17,593,495 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -33.37 |
| Energy contribution | -33.70 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17593385 110 + 22224390 CCCAGAAGCGACAAAAUUGUAACAGUGGCAAAUGCCCCAGCCCCUAAAAGCCAAAGGGGGCGUGGCCGGCAGCUGGACCAAGUGCUGCAAAAAAGGGCGAAACCCAUUGG .((((..(((.((...(((...((((.((....(((...((((((..........))))))..)))..)).))))...))).)).)))......(((.....))).)))) ( -36.20) >DroSec_CAF1 12765 104 + 1 -----AAGCGACAAAAUUGUAACAGUGGCAAAUGCCCCAGCACCUAAAAGCCAAAGGGGGCGUGGCCGGCAGCUGGACCAAGUGCUGCAAA-AAGGGCGAAACCCAUUGG -----..((((.....))))..((((((....(((((((((((......(((......))).(((((((...)))).))).))))))....-..)))))....)))))). ( -35.00) >DroSim_CAF1 31404 104 + 1 -----AAGCGACAAAAUUGUAACAGUGGCAAAUGCCCCAGCCCCUAAAAGCCAAAGGGGGCGUGGCUGGCAGCUGGACCAAGUGCUGCAAA-AAGGGCGAAACCCAUUGG -----..((((.....))))..((((((....(((((((((((((..........)))))).))....(((((..........)))))...-..)))))....)))))). ( -35.90) >consensus _____AAGCGACAAAAUUGUAACAGUGGCAAAUGCCCCAGCCCCUAAAAGCCAAAGGGGGCGUGGCCGGCAGCUGGACCAAGUGCUGCAAA_AAGGGCGAAACCCAUUGG .......((((.....))))..((((((....(((((((((((((..........)))))).))....(((((..........)))))......)))))....)))))). (-33.37 = -33.70 + 0.33)


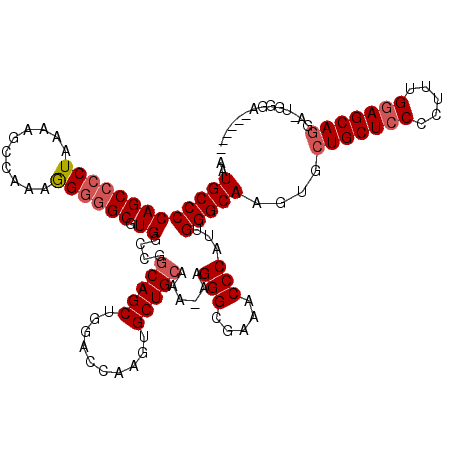
| Location | 17,593,415 – 17,593,528 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.10 |
| Mean single sequence MFE | -44.98 |
| Consensus MFE | -37.64 |
| Energy contribution | -38.48 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17593415 113 + 22224390 AAUGCCCCAGCCCCUAAAAGCCAAAGGGGGCGUGGCCGGCAGCUGGACCAAGUGCUGCAAAAAAGGGCGAAACCCAUUGGGCAAAUGCUGCUCCCCUUUGAAGCAGGA-UGGGA------ .....((((...(((....((((((((((((((.(((.(((((..........)))))......(((.....)))....)))....)).)).))))))))..))))).-)))).------ ( -42.70) >DroSec_CAF1 12790 112 + 1 AAUGCCCCAGCACCUAAAAGCCAAAGGGGGCGUGGCCGGCAGCUGGACCAAGUGCUGCAAA-AAGGGCGAAACCCAUUGGGCAAGUGCUGCUCCCCUUUGGAGCAGGA-UUGGA------ ..(((((((((((......(((......))).(((((((...)))).))).))))))....-..(((.....)))...)))))(((.(((((((.....))))))).)-))...------ ( -47.20) >DroSim_CAF1 31429 112 + 1 AAUGCCCCAGCCCCUAAAAGCCAAAGGGGGCGUGGCUGGCAGCUGGACCAAGUGCUGCAAA-AAGGGCGAAACCCAUUGGGCAAGUGCUGCUCCCCUUUGGAGCAGGA-UGGGA------ ..(((((((((((((..........)))))).))....(((((..........)))))...-..)))))...((((((.(((....)))(((((.....)))))..))-)))).------ ( -47.10) >DroEre_CAF1 12457 105 + 1 AAUGCCCCAGCC-CUAAAAAGC-AAGGGGGCGUGGCCGGCAGCUGGACCAAGUGCUGCAAA-AAGGGCGAAACCCACUGGGCAAGUGGUGCUCCCCUUUGGAGCAGGA------------ .(((((((.((.-.......))-...)))))))((((((...)))).)).....((((..(-(((((.((...(((((.....)))))...))))))))...))))..------------ ( -41.90) >DroYak_CAF1 23866 119 + 1 AAUGCCCCAGCCCCUAAAAAGCGAAAGGGGCGUGGUCGGCAGCUGGACCAAGUGCUGGAAA-AAGGGCGAAACCCAUUGGGCAAGUGCUGCUCCCCUUUGGAGCAGGAAUGGGAAUAGGG ..(((((((((((((..........)))))).)).((((((.((......))))))))...-..)))))...((((((.(((....)))(((((.....)))))...))))))....... ( -46.00) >consensus AAUGCCCCAGCCCCUAAAAGCCAAAGGGGGCGUGGCCGGCAGCUGGACCAAGUGCUGCAAA_AAGGGCGAAACCCAUUGGGCAAGUGCUGCUCCCCUUUGGAGCAGGA_UGGGA______ ..(((((((((((((..........)))))).))....(((((..........)))))......(((.....)))...)))))....(((((((.....))))))).............. (-37.64 = -38.48 + 0.84)


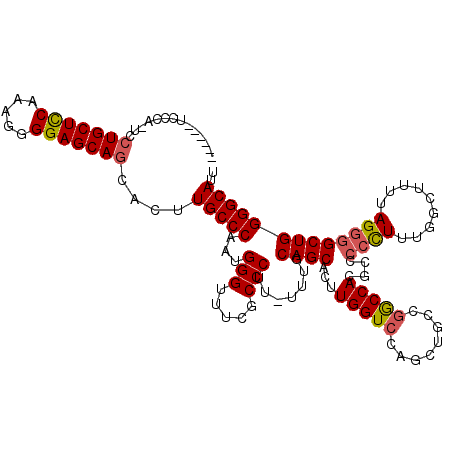
| Location | 17,593,415 – 17,593,528 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.10 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -36.76 |
| Energy contribution | -37.08 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17593415 113 - 22224390 ------UCCCA-UCCUGCUUCAAAGGGGAGCAGCAUUUGCCCAAUGGGUUUCGCCCUUUUUUGCAGCACUUGGUCCAGCUGCCGGCCACGCCCCCUUUGGCUUUUAGGGGCUGGGGCAUU ------(((((-((((...(((((((((.((.((....))((...(((.....)))......(((((..........))))).))....)))))))))))......)))).))))).... ( -41.40) >DroSec_CAF1 12790 112 - 1 ------UCCAA-UCCUGCUCCAAAGGGGAGCAGCACUUGCCCAAUGGGUUUCGCCCUU-UUUGCAGCACUUGGUCCAGCUGCCGGCCACGCCCCCUUUGGCUUUUAGGUGCUGGGGCAUU ------.....-..(((((((.....)))))))....(((((...(((.....)))..-....((((((((((((........))))..(((......)))....))))))))))))).. ( -45.60) >DroSim_CAF1 31429 112 - 1 ------UCCCA-UCCUGCUCCAAAGGGGAGCAGCACUUGCCCAAUGGGUUUCGCCCUU-UUUGCAGCACUUGGUCCAGCUGCCAGCCACGCCCCCUUUGGCUUUUAGGGGCUGGGGCAUU ------.....-..(((((((.....)))))))....(((((...(((.....)))..-...(((((..........)))))(((((..(((......))).......)))))))))).. ( -43.00) >DroEre_CAF1 12457 105 - 1 ------------UCCUGCUCCAAAGGGGAGCACCACUUGCCCAGUGGGUUUCGCCCUU-UUUGCAGCACUUGGUCCAGCUGCCGGCCACGCCCCCUU-GCUUUUUAG-GGCUGGGGCAUU ------------..((((...(((((((((..(((((.....)))))..))).)))))-)..))))....(((((........))))).(((((...-((((....)-))).)))))... ( -41.60) >DroYak_CAF1 23866 119 - 1 CCCUAUUCCCAUUCCUGCUCCAAAGGGGAGCAGCACUUGCCCAAUGGGUUUCGCCCUU-UUUCCAGCACUUGGUCCAGCUGCCGACCACGCCCCUUUCGCUUUUUAGGGGCUGGGGCAUU .((((((..((...(((((((.....)))))))....))...))))))....(((((.-...........(((((........))))).((((((..........)))))).)))))... ( -42.90) >consensus ______UCCCA_UCCUGCUCCAAAGGGGAGCAGCACUUGCCCAAUGGGUUUCGCCCUU_UUUGCAGCACUUGGUCCAGCUGCCGGCCACGCCCCCUUUGGCUUUUAGGGGCUGGGGCAUU ..............(((((((.....)))))))....(((((...(((.....))).......((((...(((((........)))))....((((.........))))))))))))).. (-36.76 = -37.08 + 0.32)

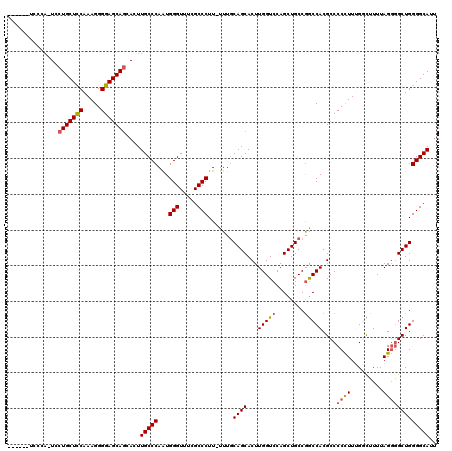
| Location | 17,593,455 – 17,593,549 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -24.41 |
| Consensus MFE | -18.84 |
| Energy contribution | -19.12 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17593455 94 - 22224390 GAUCCCGAUCCCCAUUCGCAU-------------------------UCCCA-UCCUGCUUCAAAGGGGAGCAGCAUUUGCCCAAUGGGUUUCGCCCUUUUUUGCAGCACUUGGUCCAGCU ((((..((..((((((.(((.-------------------------....(-(.(((((((.....))))))).)).)))..))))))..))((........)).......))))..... ( -24.00) >DroSec_CAF1 12830 90 - 1 ---CCCGAUCCCCAUUCGCAU-------------------------UCCAA-UCCUGCUCCAAAGGGGAGCAGCACUUGCCCAAUGGGUUUCGCCCUU-UUUGCAGCACUUGGUCCAGCU ---.((((..((((((.(((.-------------------------.....-..(((((((.....)))))))....)))..))))))..))((....-...)).......))....... ( -25.60) >DroSim_CAF1 31469 90 - 1 ---CCCGAUCCCCAUUCGCAU-------------------------UCCCA-UCCUGCUCCAAAGGGGAGCAGCACUUGCCCAAUGGGUUUCGCCCUU-UUUGCAGCACUUGGUCCAGCU ---.((((..((((((.(((.-------------------------.....-..(((((((.....)))))))....)))..))))))..))((....-...)).......))....... ( -25.60) >DroEre_CAF1 12495 78 - 1 ---U------CCAAUUCGCA--------------------------------UCCUGCUCCAAAGGGGAGCACCACUUGCCCAGUGGGUUUCGCCCUU-UUUGCAGCACUUGGUCCAGCU ---.------.......((.--------------------------------....(((((.....)))))((((..(((.(((.(((.....)))..-.)))..)))..))))...)). ( -23.40) >DroYak_CAF1 23906 116 - 1 ---CCCGAUCCCAAUUCCCAUUCCAGAUCCCAUUCCCGCUCCCUAUUCCCAUUCCUGCUCCAAAGGGGAGCAGCACUUGCCCAAUGGGUUUCGCCCUU-UUUCCAGCACUUGGUCCAGCU ---.......((((...........((.((((((...(((((((....................))))))).((....))..))))))..))((....-......))..))))....... ( -23.45) >consensus ___CCCGAUCCCCAUUCGCAU_________________________UCCCA_UCCUGCUCCAAAGGGGAGCAGCACUUGCCCAAUGGGUUUCGCCCUU_UUUGCAGCACUUGGUCCAGCU .................((...................................(((((((.....)))))))((..(((.(((.(((.....)))....)))..)))..)).....)). (-18.84 = -19.12 + 0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:31 2006