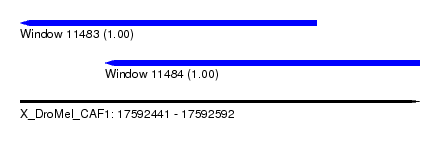
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,592,441 – 17,592,592 |
| Length | 151 |
| Max. P | 0.999945 |

| Location | 17,592,441 – 17,592,553 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.88 |
| Mean single sequence MFE | -36.04 |
| Consensus MFE | -34.60 |
| Energy contribution | -34.64 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.26 |
| SVM RNA-class probability | 0.999853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17592441 112 - 22224390 GUGCGAACGCAUUUUGGCUCCCAAGACGAGUUCGUUACGCGCGUAAUUGCGAAAGUAAUUAGUGCAAAGCGUUCAAUUGCGAUACUUGCAACCUGUAACUGGUUAAAUUCCC-------- ....((((((...((((...))))((((....))))..((((.(((((((....)))))))))))...))))))....((((...))))((((.......))))........-------- ( -33.70) >DroSec_CAF1 11832 112 - 1 GUGCGAACGCAUUUUGGCUCCCAAGACGAGUUCGUUACGCGCGUAAUUGCGAAAGUAAUUAGUGCAAAGCGUUCAAUUGCGAUACUCGCAACCUGUAACUGGUUAAAUCCCC-------- ....((((((...((((...))))((((....))))..((((.(((((((....)))))))))))...))))))....(((.....)))((((.......))))........-------- ( -36.10) >DroSim_CAF1 30452 112 - 1 GUGCGAACGCAUUUUGGCUCCCAAGACGAGUUCGUUACGCGCGUAAUUGCGAAAGUAAUUAGUGCAAAGCGUUCAAUUGCGAUACUCGCAACCUGUAACUGGUUAAAUCCCC-------- ....((((((...((((...))))((((....))))..((((.(((((((....)))))))))))...))))))....(((.....)))((((.......))))........-------- ( -36.10) >DroEre_CAF1 11578 112 - 1 GUGCGAACGCAUUUUGGCUCCCAAGACGAGUUCGUUACGCGCGUAAUUGCGAAAGUAAUUAGUGCAAAGCGUUCAAUUGCGAUUCUCGCCACCUGUAACUGGUUAAAGCCCC-------- (((.((((((...((((...))))((((....))))..((((.(((((((....)))))))))))...))))))....(((.....))))))........(((....)))..-------- ( -35.00) >DroYak_CAF1 22830 120 - 1 GUGCGAACGCAUUUUGGCUCCCAAGACGAGUUCGUUACGCGCGUAAUUGCGAAAGUAAUUAGUGCAAAGCGUUCAAUUGCGAUACUCGCAACCUGUAACUGCUUAAAACCCCCCCACUUC ((((((((.(.((((((...)))))).).))))).)))((((.(((((((....))))))))))).(((((((((.(((((.....)))))..)).))).))))................ ( -39.30) >consensus GUGCGAACGCAUUUUGGCUCCCAAGACGAGUUCGUUACGCGCGUAAUUGCGAAAGUAAUUAGUGCAAAGCGUUCAAUUGCGAUACUCGCAACCUGUAACUGGUUAAAUCCCC________ .(((((((((...((((...))))((((....))))..((((.(((((((....)))))))))))...))))))..(((((.....)))))...)))....................... (-34.60 = -34.64 + 0.04)

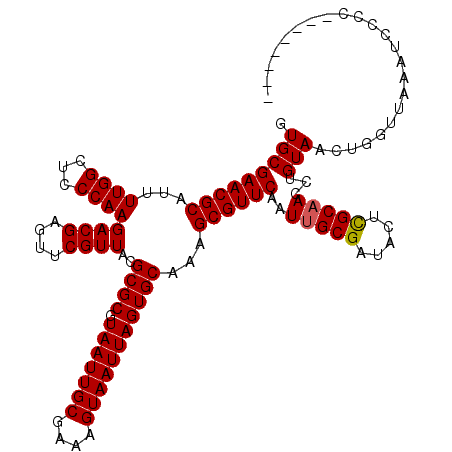

| Location | 17,592,473 – 17,592,592 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -42.02 |
| Consensus MFE | -41.74 |
| Energy contribution | -41.74 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.74 |
| SVM RNA-class probability | 0.999945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17592473 119 - 22224390 GGAGGGUGGAGCACACAAAA-GGGGGCCUAAUGAUGGGAAGUGCGAACGCAUUUUGGCUCCCAAGACGAGUUCGUUACGCGCGUAAUUGCGAAAGUAAUUAGUGCAAAGCGUUCAAUUGC ........((((.(......-(((((((.........(((((((....))))))))))))))..((((....))))..((((.(((((((....)))))))))))...).))))...... ( -41.90) >DroSec_CAF1 11864 119 - 1 GGAGGAUGGAGCACACAAAA-GGGGGCCUAAUGAUGGGAAGUGCGAACGCAUUUUGGCUCCCAAGACGAGUUCGUUACGCGCGUAAUUGCGAAAGUAAUUAGUGCAAAGCGUUCAAUUGC ....(((.((((.(......-(((((((.........(((((((....))))))))))))))..((((....))))..((((.(((((((....)))))))))))...).)))).))).. ( -42.10) >DroSim_CAF1 30484 119 - 1 GGAGGGUGGAGCACACAAAA-GGGGGCCUAAUGAUGGGAAGUGCGAACGCAUUUUGGCUCCCAAGACGAGUUCGUUACGCGCGUAAUUGCGAAAGUAAUUAGUGCAAAGCGUUCAAUUGC ........((((.(......-(((((((.........(((((((....))))))))))))))..((((....))))..((((.(((((((....)))))))))))...).))))...... ( -41.90) >DroEre_CAF1 11610 120 - 1 CAAGGGUGGAGCACACAAAAAGGGGGCCUAAUGAUGGGAAGUGCGAACGCAUUUUGGCUCCCAAGACGAGUUCGUUACGCGCGUAAUUGCGAAAGUAAUUAGUGCAAAGCGUUCAAUUGC (((...((((((.........(((((((.........(((((((....))))))))))))))..((((....))))..((((.(((((((....)))))))))))...)).)))).))). ( -42.40) >DroYak_CAF1 22870 120 - 1 AGGGGGUGGAGCACACAAAAGGGGGGCCUAAUGAUGGGAAGUGCGAACGCAUUUUGGCUCCCAAGACGAGUUCGUUACGCGCGUAAUUGCGAAAGUAAUUAGUGCAAAGCGUUCAAUUGC ..(.(((.((((.(.......(((((((.........(((((((....))))))))))))))..((((....))))..((((.(((((((....)))))))))))...).)))).))).) ( -41.80) >consensus GGAGGGUGGAGCACACAAAA_GGGGGCCUAAUGAUGGGAAGUGCGAACGCAUUUUGGCUCCCAAGACGAGUUCGUUACGCGCGUAAUUGCGAAAGUAAUUAGUGCAAAGCGUUCAAUUGC ........((((.(.......(((((((.........(((((((....))))))))))))))..((((....))))..((((.(((((((....)))))))))))...).))))...... (-41.74 = -41.74 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:28 2006