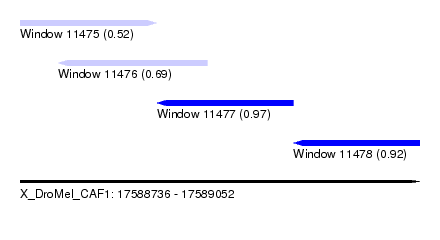
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,588,736 – 17,589,052 |
| Length | 316 |
| Max. P | 0.967889 |

| Location | 17,588,736 – 17,588,844 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.41 |
| Mean single sequence MFE | -51.19 |
| Consensus MFE | -32.88 |
| Energy contribution | -33.63 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17588736 108 + 22224390 ACCCCC---UAA-------CCACCCAGCCUGUCAGUUCCACGCCGCCCCUGGCAGCCCCCCCCCCUUUCGGACG--AGCGGUGGAAGGGGGGGGAGGGGGUAUCACGGCUGGGGCCUCGU ......---...-------...(((((((((..(.((((..((.(((...))).)).(((((((((((((..(.--...).))))))))))))).)))).)..)).)))))))....... ( -53.20) >DroSec_CAF1 8160 102 + 1 ACUCCC---UUA-GACCCCCUACCCAGCCUGUCAGUUCCACGCCGCCCCUGGCAGCCCCCCC--CUUUCGCA-G-----------AGCGGGGGGAGGGGGUAUCACGGCUGGGGCCUCGU ......---...-.........(((((((((((((.............))))))((((((.(--((..(((.-.-----------.)))..))).)))))).....)))))))....... ( -46.02) >DroSim_CAF1 27077 101 + 1 ACCCCC---UUA-GACCCCCUACCCAGCCUGUCAGUUCCACGCCGCCCCUGGCAGCCCCCCC--C-UUCGCA-G-----------AGCGGGGGGAGGGGGUAUCACGGCUGGGGCCUCGU ......---...-.........(((((((((((((.............))))))((((((((--(-(((((.-.-----------.)))))))).)))))).....)))))))....... ( -47.62) >DroYak_CAF1 18659 113 + 1 ACUCCCCCCUUCCCCCCCCCUGCCCAGCCUGUCAGUUGCACGCCGCCCCUGGCAGCCCAC----CUUUCGGA-GAGAGGGGUGGA--GGGGCGGAGGGGGUAUCACGGCUGGGGCCUCGU .....((((..(((((((.((((((...(((((((..((.....))..))))))).((((----(((((...-..))))))))).--.)))))).)))))......))..))))...... ( -57.90) >consensus ACCCCC___UUA_GACCCCCUACCCAGCCUGUCAGUUCCACGCCGCCCCUGGCAGCCCCCCC__CUUUCGCA_G___________AGCGGGGGGAGGGGGUAUCACGGCUGGGGCCUCGU ......................(((((((.....(.....)...((((((.....((((((...........................)))))).)))))).....)))))))....... (-32.88 = -33.63 + 0.75)


| Location | 17,588,766 – 17,588,884 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -36.98 |
| Consensus MFE | -26.80 |
| Energy contribution | -26.92 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17588766 118 - 22224390 AAUUACGCUUUAACUUGUAAUAAAGAAGACAAACAAUAAAACGAGGCCCCAGCCGUGAUACCCCCUCCCCCCCCUUCCACCGCU--CGUCCGAAAGGGGGGGGGGCUGCCAGGGGCGGCG .....((((....(((((......................)))))((((((((((.....).....((((((((((.(......--.....).))))))))))))))....))))))))) ( -41.15) >DroSec_CAF1 8196 106 - 1 AAUUACGCUUGAACUUGUAAUAAAGAAGACAAACAAUAAAACGAGGCCCCAGCCGUGAUACCCCCUCCCCCCGCU-----------C-UGCGAAAG--GGGGGGGCUGCCAGGGGCGGCG .....((((....(((((......................)))))(((((.((.(.....((((((((...(((.-----------.-.)))...)--)))))))).))..))))))))) ( -35.75) >DroSim_CAF1 27113 105 - 1 AAUUACACUUUAACUUGUAAUAAAGAAGACAAACAAUAAAACGAGGCCCCAGCCGUGAUACCCCCUCCCCCCGCU-----------C-UGCGAA-G--GGGGGGGCUGCCAGGGGCGGCG .(((((.(((((........)))))...................(((....))))))))...(((((((..(((.-----------.-.)))..-)--))))))((((((...)))))). ( -36.10) >DroYak_CAF1 18699 113 - 1 AAUUACGCUUUAACUUGUAAUAAAGAAGACAAACAAUAAAACGAGGCCCCAGCCGUGAUACCCCCUCCGCCCC--UCCACCCCUCUC-UCCGAAAG----GUGGGCUGCCAGGGGCGGCG .(((((.(((((........)))))...................(((....)))))))).......(((((((--(.((.(((....-.((....)----).))).))..)))))))).. ( -34.90) >consensus AAUUACGCUUUAACUUGUAAUAAAGAAGACAAACAAUAAAACGAGGCCCCAGCCGUGAUACCCCCUCCCCCCCCU___________C_UCCGAAAG__GGGGGGGCUGCCAGGGGCGGCG .(((((.......(((......)))...................(((....)))))))).......((((((..........................))))))((((((...)))))). (-26.80 = -26.92 + 0.12)

| Location | 17,588,844 – 17,588,952 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 98.15 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -24.88 |
| Energy contribution | -24.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17588844 108 - 22224390 CUUCAAGUUUGUGGCUCAAGUUGCCUGAACUUGCUGUUCGCUUGCAAAUUGCCGCAUAUUUGUUGAGUAAUUACGCUUUAACUUGUAAUAAAGAAGACAAACAAUAAA ((((.....((((((.....((((.(((((.....)))))...))))...))))))(((..(((((((......)).)))))..))).....))))............ ( -25.00) >DroSec_CAF1 8262 108 - 1 CUUCAAGUUUGUGGCUCAAGUUGCCUGAACUUGCUGUUCGCUUGCAAAUUGCCGCAUAUUUGUUGAGUAAUUACGCUUGAACUUGUAAUAAAGAAGACAAACAAUAAA ((((((((.((((((.....((((.(((((.....)))))...))))...)))))).)))).(..(((......)))..)............))))............ ( -25.40) >DroSim_CAF1 27178 108 - 1 CUUCAAGUUUGUGGCUCAAGUUGCCUGAACUUGCUGUUCGCUUGCAAAUUGCCGCAUAUUUGUUGAGUAAUUACACUUUAACUUGUAAUAAAGAAGACAAACAAUAAA ((((((((.((((((.....((((.(((((.....)))))...))))...)))))).))))........((((((........))))))...))))............ ( -25.20) >DroEre_CAF1 8170 108 - 1 CUUCGAGCUUGUGGCUCAACUUGCCUGAACUUGCUGUUCGCUUGCAAAUUGCCGCAUAUUUGUUGAGUAAUUACGCUUUAACUUGUAAUAAAGAAGACAAACAAUAAA ((((.....((((((.....((((.(((((.....)))))...))))...))))))(((..(((((((......)).)))))..))).....))))............ ( -23.90) >DroYak_CAF1 18772 108 - 1 CUUCAAGUUUGUGGCUCAAGUUGCCUGAACUUGCUGUUCGCUUGCAAAUUGCCGCAUAUUUGUUGAGUAAUUACGCUUUAACUUGUAAUAAAGAAGACAAACAAUAAA ((((.....((((((.....((((.(((((.....)))))...))))...))))))(((..(((((((......)).)))))..))).....))))............ ( -25.00) >consensus CUUCAAGUUUGUGGCUCAAGUUGCCUGAACUUGCUGUUCGCUUGCAAAUUGCCGCAUAUUUGUUGAGUAAUUACGCUUUAACUUGUAAUAAAGAAGACAAACAAUAAA ((((.....((((((.....((((.(((((.....)))))...))))...))))))(((..(((((((......)))).)))..))).....))))............ (-24.88 = -24.72 + -0.16)


| Location | 17,588,952 – 17,589,052 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.09 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -10.11 |
| Energy contribution | -11.05 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.35 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17588952 100 - 22224390 CGGGCGGAGCCUUCCCUUUGGUUUCUUCGGAUCCCUCACU-UCUG----------AAGUAAGCCCCACACCCCCUUCCUGUUUCUACGGGCGGUAUGCAUCCCAUUCGAUU .((((((((((........))))))((((((.........-))))----------))....))))((.((((((.............))).))).)).............. ( -27.12) >DroSec_CAF1 8370 100 - 1 CGGGCGGAGCCUUCCCUUUGGUUUCUUCGGACCCCUCACU-UCUG----------AAGAAACCCCCACAUCCCCUUCAUCCUUCUACAUGGGGCAUCCAUCCAAUUCGAAU .((..((((((........((((((((((((.........-))))----------))))))))................((........))))).)))..))......... ( -29.90) >DroSim_CAF1 27286 100 - 1 CGGGCGGAGCCUUCCCUUUAGUUUCUUCGGACCCCUCACU-UCUG----------AAGAAACCCCCACACCCGCUUCAUCCUUCUACAUGGGGCAUCCAUCCAAUUCGAAU .((..(((............(((((((((((.........-))))----------)))))))..........(((((((........))))))).)))..))......... ( -27.30) >DroEre_CAF1 8278 98 - 1 CGGACGGAGCCUUCCCCAUGGUUUCUUCGGACUCCUCUCCCGCU-CCCUCCUCUGGAGAGCCCCCCGCACU------------CCAUACCCCCUGCCCAUCCAAUGGGAAU .(((.(((((.........((..((....))..))......)))-)).)))..(((((.((.....)).))------------))).........(((((...)))))... ( -30.96) >consensus CGGGCGGAGCCUUCCCUUUGGUUUCUUCGGACCCCUCACU_UCUG__________AAGAAACCCCCACACCCCCUUCAUCCUUCUACAUGCGGCAUCCAUCCAAUUCGAAU .(((.((((((........((..((....))..))............................................((........))))).))).)))......... (-10.11 = -11.05 + 0.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:22 2006