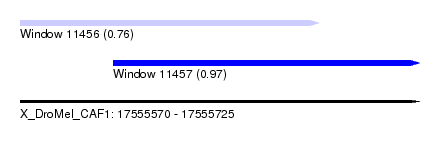
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,555,570 – 17,555,725 |
| Length | 155 |
| Max. P | 0.965872 |

| Location | 17,555,570 – 17,555,686 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.39 |
| Mean single sequence MFE | -20.30 |
| Consensus MFE | -14.65 |
| Energy contribution | -14.77 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
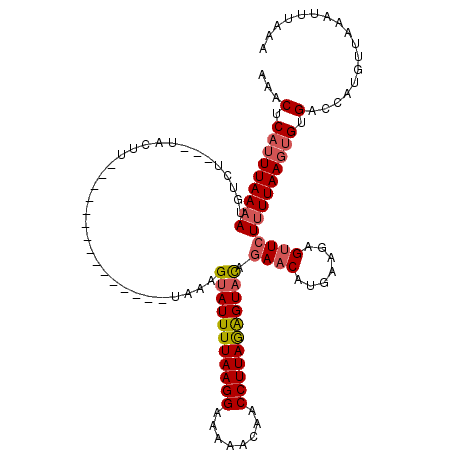
>X_DroMel_CAF1 17555570 116 + 22224390 AUUUCAAAAAAGAUGAAACUGUUGUGUUGUUUUGCAAAACUCUAUUAAAAUGUCACCCUACCUUUGUCUAUUUUUGGCUAAAGUAUUUUAAGGAAAAACAACCUUAGAGUUUAGAA ...((((((((((..((..((..(((.(((((((...........))))))).)))..))..))..))).)))))))((((...(((((((((........))))))))))))).. ( -19.30) >DroSec_CAF1 7482 98 + 1 AUUAAAAAGAAGGUGAAGCUGUUGCGUUGUUUUGCAAAACUCAUUUAAAAUGGCU---UACUU---------------UAAAGUAUUUUAAGGAAAAACAACCUUAAAGUACAGAU .........(((((((.((..(((((......)))))....(((.....))))))---)))))---------------)...(((((((((((........))))))))))).... ( -22.60) >DroSim_CAF1 7305 98 + 1 AUUAAAAAGAAGGUGAAGCUGUUGUGUUGUUUUGCAAAACUCAUUUAAAAUGUCU---UACUU---------------UAAAGUAUUUUAAGGAAAAACAACCUUAGGGUACAGAA .........(((((((..(....(((..((((....)))).))).......)..)---)))))---------------)...(((((((((((........))))))))))).... ( -19.00) >consensus AUUAAAAAGAAGGUGAAGCUGUUGUGUUGUUUUGCAAAACUCAUUUAAAAUGUCU___UACUU_______________UAAAGUAUUUUAAGGAAAAACAACCUUAGAGUACAGAA ..........((((((.....(((((......)))))...))))))....................................(((((((((((........))))))))))).... (-14.65 = -14.77 + 0.12)


| Location | 17,555,606 – 17,555,725 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -21.60 |
| Consensus MFE | -13.83 |
| Energy contribution | -15.17 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17555606 119 + 22224390 AAACUCUAUUAAAAUGUCACCCUACCUUUGUCUAUUUUUGGCUAAAGUAUUUUAAGGAAAAACAACCUUAGAGUUUAGAACAUAAAGAGUUAU-UUAAGUGUGACCAAGUUAAAUUAAAA .((((((..................(((((.(((....))).))))).(((((((((........)))))))))...........)))))).(-((((...((((...))))..))))). ( -18.10) >DroSec_CAF1 7518 102 + 1 AAACUCAUUUAAAAUGGCU---UACUU---------------UAAAGUAUUUUAAGGAAAAACAACCUUAAAGUACAGAUCAUGAAGAGUUCUUUUAAGUGUGACCAUGUUAAAUUUAAA ......((((((.((((.(---(((((---------------(((((((((((((((........)))))))))))(((.(.......).))))))))).)))))))).))))))..... ( -22.80) >DroSim_CAF1 7341 102 + 1 AAACUCAUUUAAAAUGUCU---UACUU---------------UAAAGUAUUUUAAGGAAAAACAACCUUAGGGUACAGAACAUGAAGUGUUCUUUUAAGUGUGACCAUAUUAAAUUUAAA ......((((((.(((((.---(((((---------------.((((((((((((((........)))))))))))(((((((...))))))))))))))).)).))).))))))..... ( -23.90) >consensus AAACUCAUUUAAAAUGUCU___UACUU_______________UAAAGUAUUUUAAGGAAAAACAACCUUAGAGUACAGAACAUGAAGAGUUCUUUUAAGUGUGACCAUGUUAAAUUUAAA ...(.(((((((((................................(((((((((((........))))))))))).((((.......))))))))))))).)................. (-13.83 = -15.17 + 1.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:03 2006