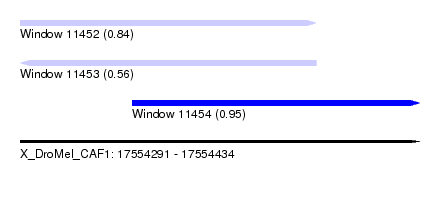
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,554,291 – 17,554,434 |
| Length | 143 |
| Max. P | 0.954791 |

| Location | 17,554,291 – 17,554,397 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 87.42 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -18.42 |
| Energy contribution | -19.67 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17554291 106 + 22224390 UGAGUUGCGUGUAGAUAACACGACUAUGUCUGCUAGAUUGUAAACUUACACGAAUUUUACAUUUGCUAUAGGAAAUAAUGCCUAUAAGUAGGCAGCACUUUUCCAA .((((.((((((.....)))).....((((((((.((.((((((.((.....)).)))))).))..((((((........))))))))))))))))))))...... ( -25.50) >DroSec_CAF1 6195 106 + 1 UGAGUUGCGUGUAGAUAACACGACCAUGUCUAGAAAGAUGUAAACUUACACGCAUUUUACAUUUGCUAUAGGAAAUAAUGCCUAUAAGUAGGAAGCACUUUUCCAA ((((.(((((((((......)....((((((....)))))).....)))))))).))))....(((((((((........))))).))))(((((....))))).. ( -23.60) >DroSim_CAF1 6030 106 + 1 UACGUGGCGUGUAGAUAACACGACCAUGUCUAGAAAGAUGUAAACUUACAGGAAUUUUACAUUUGCUAUAGGAAAUAAUGCCUAUAAGUAGGCAGCACUUUUCCAA .(((((((((((.....))))).)))))).(((..(((((((((.((.....)).))))))))).)))..(((((...((((((....)))))).....))))).. ( -33.00) >DroEre_CAF1 10771 106 + 1 UAUGUUGCGUGUAGAGAACACGACCAUGUCCAUAAAUAUGUAAUUUUACGCGCAUUUUACAUUCGCUUUAGGAAAUAAUGCCUAAAAGUAGGCAGCGUUUUACCAA .(((((((((((((((.....(((...)))(((....)))...)))))))))))....)))).........(((((..((((((....))))))..)))))..... ( -23.90) >consensus UAAGUUGCGUGUAGAUAACACGACCAUGUCUAGAAAGAUGUAAACUUACACGAAUUUUACAUUUGCUAUAGGAAAUAAUGCCUAUAAGUAGGCAGCACUUUUCCAA ......((((((.....))))..............(((((((((...........)))))))))))....(((((...((((((....)))))).....))))).. (-18.42 = -19.67 + 1.25)



| Location | 17,554,291 – 17,554,397 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 87.42 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -16.02 |
| Energy contribution | -16.83 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17554291 106 - 22224390 UUGGAAAAGUGCUGCCUACUUAUAGGCAUUAUUUCCUAUAGCAAAUGUAAAAUUCGUGUAAGUUUACAAUCUAGCAGACAUAGUCGUGUUAUCUACACGCAACUCA ..((((..(((.((((((....)))))).)))))))....((.............((((..(((........)))..))))....((((.....))))))...... ( -21.30) >DroSec_CAF1 6195 106 - 1 UUGGAAAAGUGCUUCCUACUUAUAGGCAUUAUUUCCUAUAGCAAAUGUAAAAUGCGUGUAAGUUUACAUCUUUCUAGACAUGGUCGUGUUAUCUACACGCAACUCA ..((((((((((((.........))))))).)))))................((((((((..(..((((....(((....)))..)))).)..))))))))..... ( -24.30) >DroSim_CAF1 6030 106 - 1 UUGGAAAAGUGCUGCCUACUUAUAGGCAUUAUUUCCUAUAGCAAAUGUAAAAUUCCUGUAAGUUUACAUCUUUCUAGACAUGGUCGUGUUAUCUACACGCCACGUA (((((((..((......(((((((((....((((..((((.....)))))))).)))))))))...))..)))))))((.(((.(((((.....)))))))).)). ( -26.30) >DroEre_CAF1 10771 106 - 1 UUGGUAAAACGCUGCCUACUUUUAGGCAUUAUUUCCUAAAGCGAAUGUAAAAUGCGCGUAAAAUUACAUAUUUAUGGACAUGGUCGUGUUCUCUACACGCAACAUA .........(((((((((....))))).(((.....))))))).((((....((((.(((((((.....))))..((((((....))))))..))).)))))))). ( -22.10) >consensus UUGGAAAAGUGCUGCCUACUUAUAGGCAUUAUUUCCUAUAGCAAAUGUAAAAUGCGUGUAAGUUUACAUCUUUCUAGACAUGGUCGUGUUAUCUACACGCAACUCA ..((((..(((.((((((....)))))).)))))))....((..((((........((((....)))).........))))....((((.....))))))...... (-16.02 = -16.83 + 0.81)

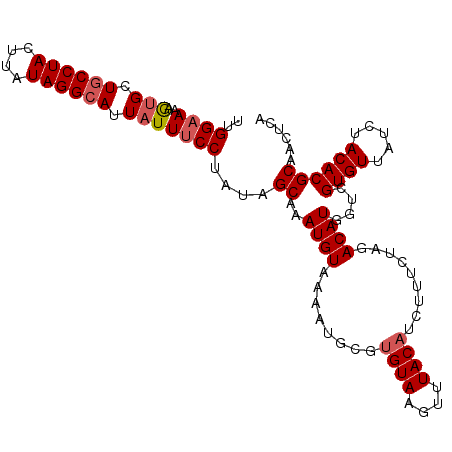
| Location | 17,554,331 – 17,554,434 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.63 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -12.83 |
| Energy contribution | -13.03 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17554331 103 + 22224390 UAAACUUACACGAAUUUUACAUUUGCUAUAGGAAAUAAUGCCUAUAAGUAGGCAGCACUUUUC-CAA---UAGCUGCUUAAGGCAUUCGCCAACAGGUGGCGCCACU ........................(((((.(((((...((((((....)))))).....))))-).)---)))).......(((..(((((....))))).)))... ( -30.00) >DroSec_CAF1 6235 103 + 1 UAAACUUACACGCAUUUUACAUUUGCUAUAGGAAAUAAUGCCUAUAAGUAGGAAGCACUUUUC-CAA---UAGCUGCUUAAGACAUUCGCCCACAGGUGGCGCCACU ..........(((.(((((.....(((((.(((((.....((((....)))).......))))-).)---))))....)))))....((((....)))))))..... ( -22.70) >DroSim_CAF1 6070 103 + 1 UAAACUUACAGGAAUUUUACAUUUGCUAUAGGAAAUAAUGCCUAUAAGUAGGCAGCACUUUUC-CAA---UAGCUGCUUAAGACAUUCGCCAACAGGUGGCGCCACU ..........((..(((((.....(((((.(((((...((((((....)))))).....))))-).)---))))....)))))....(((((.....)))))))... ( -28.40) >DroEre_CAF1 10811 103 + 1 UAAUUUUACGCGCAUUUUACAUUCGCUUUAGGAAAUAAUGCCUAAAAGUAGGCAGCGUUUUAC-CAA---AUGCCGCUUAAGGCGCUUGCCAACAGGUGGCGCCACU .........((((...........((((((((......((((((....))))))((((((...-.))---))))..))))))))(((((....))))).)))).... ( -31.40) >DroAna_CAF1 12806 99 + 1 ----UUUACAUGGUUUCUACAUUUG-UGUAAAAAGUAAAGCCUACAAGCAGGCAGUGGUUUUUGUAAAUGUAGUUUCUUU---UGCUCAGCGUUAGAUGGCGCCACA ----......((((..(((((((((-..((((((.((..((((......))))..)).)))))))))))))))..(((..---(((...)))..)))....)))).. ( -24.00) >consensus UAAACUUACACGAAUUUUACAUUUGCUAUAGGAAAUAAUGCCUAUAAGUAGGCAGCACUUUUC_CAA___UAGCUGCUUAAGACAUUCGCCAACAGGUGGCGCCACU ...................((((((.((((((........))))))..((((((((................)))))))).............))))))........ (-12.83 = -13.03 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:28:01 2006