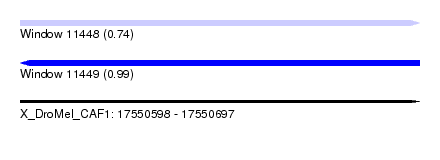
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,550,598 – 17,550,697 |
| Length | 99 |
| Max. P | 0.988567 |

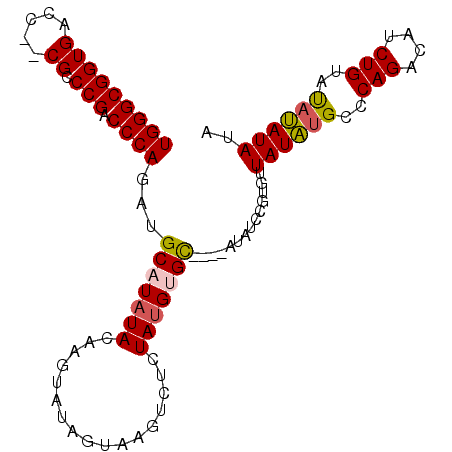
| Location | 17,550,598 – 17,550,697 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -14.69 |
| Energy contribution | -13.88 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17550598 99 + 22224390 UAUAUAUAUACAGAUGUCUGCGCAUAUAACACGGAUAUAGAGACACAUAGAGACUUACUAUACUCGUAUAUGCAUCUGGGUCGGGCG--GGUCACCGCCCA ..........((((((....)((((((...(((..(((((.................)))))..))))))))))))))....(((((--(....)))))). ( -30.33) >DroSim_CAF1 1563 99 + 1 CAUAUAUACACAGAUGUCUGGGCAUAUAACACGGCUAUAGAGGCACAUAGAGACUUACUAUACUCGUAUAUGCAUCUGGGUCGGUCG--GGUCACCGCCCA ..........(((....))).((((((...(((..(((((((((.......).))).)))))..)))))))))....(((.((((..--....))))))). ( -26.30) >DroEre_CAF1 7150 96 + 1 UAUAUGUGUACAGAUGUCUGGGCACAUAACGCGGAUAU----GCGCAUAGAGACUUA-UGUAGAUAUAUAUGCAUCUGGGUCGGGCGGGGGUCACCGCCCA ..((((((..(((....)))..))))))..(((.....----.))).....((((((-((((........)))))..)))))((((((......)))))). ( -38.00) >DroAna_CAF1 6878 86 + 1 -CUAUAUAAACAGAUGUCUGGGCCUAUAUCC---AUAC----ACCCGUAG-----UUCUCUGUUUUUAUAUGCAUCUGGGUCGGGCG--GGUCACCGCCCA -.........(((((((..(((..(((....---))).----.))).(((-----....))).........)))))))....(((((--(....)))))). ( -26.90) >consensus UAUAUAUAUACAGAUGUCUGGGCAUAUAACACGGAUAU____ACACAUAGAGACUUACUAUACUCGUAUAUGCAUCUGGGUCGGGCG__GGUCACCGCCCA ..(((((((.(((....)))..........................((((.......))))....))))))).....(((.(((..........)))))). (-14.69 = -13.88 + -0.81)

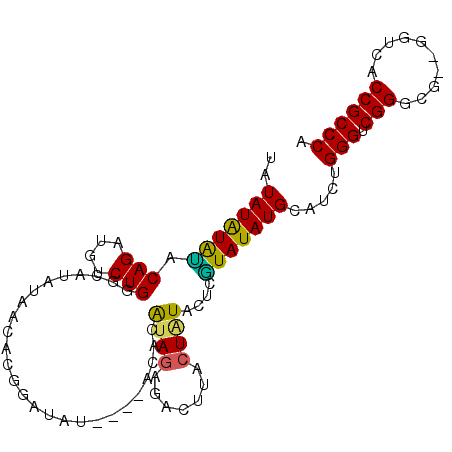
| Location | 17,550,598 – 17,550,697 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -17.17 |
| Energy contribution | -17.36 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17550598 99 - 22224390 UGGGCGGUGACC--CGCCCGACCCAGAUGCAUAUACGAGUAUAGUAAGUCUCUAUGUGUCUCUAUAUCCGUGUUAUAUGCGCAGACAUCUGUAUAUAUAUA .((((((....)--))))).....(((.(((((((.(((..........)))))))))).)))..........((((((..(((....)))..)))))).. ( -31.40) >DroSim_CAF1 1563 99 - 1 UGGGCGGUGACC--CGACCGACCCAGAUGCAUAUACGAGUAUAGUAAGUCUCUAUGUGCCUCUAUAGCCGUGUUAUAUGCCCAGACAUCUGUGUAUAUAUG .(((((((....--..)))).)))(((.(((((((.(((..........)))))))))).)))..........(((((((.(((....))).))))))).. ( -31.90) >DroEre_CAF1 7150 96 - 1 UGGGCGGUGACCCCCGCCCGACCCAGAUGCAUAUAUAUCUACA-UAAGUCUCUAUGCGC----AUAUCCGCGUUAUGUGCCCAGACAUCUGUACACAUAUA .((((((......))))))(((..(((((......)))))...-...))).....((((----......))))((((((..(((....)))..)))))).. ( -29.40) >DroAna_CAF1 6878 86 - 1 UGGGCGGUGACC--CGCCCGACCCAGAUGCAUAUAAAAACAGAGAA-----CUACGGGU----GUAU---GGAUAUAGGCCCAGACAUCUGUUUAUAUAG- .((((((....)--)))))............((((.(((((((...-----((..((((----.(((---....))).))))))...))))))).)))).- ( -29.70) >consensus UGGGCGGUGACC__CGCCCGACCCAGAUGCAUAUACAAGUAUAGUAAGUCUCUAUGUGC____AUAUCCGUGUUAUAUGCCCAGACAUCUGUAUAUAUAUA (((((((((.....)).))).))))...(((((((.................)))))))..............((((((..(((....)))..)))))).. (-17.17 = -17.36 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:56 2006