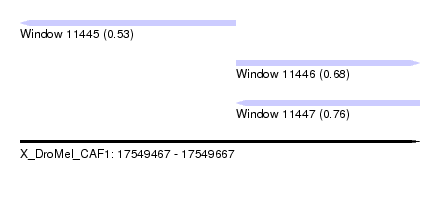
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,549,467 – 17,549,667 |
| Length | 200 |
| Max. P | 0.756857 |

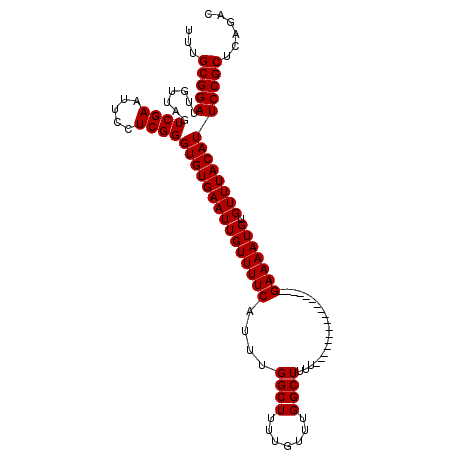
| Location | 17,549,467 – 17,549,575 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 85.96 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -23.10 |
| Energy contribution | -24.47 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.525972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17549467 108 - 22224390 UUAUUUGGAUUUGAUUCGUUUGGAUGAUGAUGAUGGUGAUGAUGGCGGAAG--AACAAGUGUUUUGCCAGCCCAAGCCCAAAUGUGAUUCAAUGCGCUGGGAAUUCAAAA ...(((((((((....(((((((((.((.((...(((.....((((((((.--(.....).))))))))......)))...)))).)))))).)))....))))))))). ( -26.20) >DroSec_CAF1 1334 99 - 1 UUAUUUGGAUUCGAUUCGUAUGGAU---------GGUGAUGAUGGCGGAAG--AACAUGUGUUUUGCCAGCCCAAGCCCAAAUGUGAUUCCAUGCGCUGGGAAUCCAGAA ...(((((((((....((((((((.---------(((.....((((((((.--(.....).))))))))......)))((....))..))))))))....))))))))). ( -33.40) >DroSim_CAF1 487 99 - 1 UUAUUUGGAUUCGAUUCGUAUGGAC---------GGUGAUGAUGGCGGAAG--AACAAGUGUUUUGCCAGCCGAAGCCCAAAUGUGAUUCCAUGCGCUGGGAAUCCAGAA ...(((((((((....((((((((.---------(((..((.((((((((.--(.....).))))))))..))..)))((....))..))))))))....))))))))). ( -34.20) >DroEre_CAF1 6004 100 - 1 UUAUUUUGAUUUGAUCCGUUUGGAU---------GG-GCUGAUGGGGGAAGUAAACAAGUGUUCUGGCAGCCCAAGCCCAAAUAUGAUUCAAUGCGCUGGGAAUCCAACA .................(((.((((---------((-((((..((((.(..(....)..).))))..))))))...((((..(((......)))...)))).))))))). ( -24.30) >consensus UUAUUUGGAUUCGAUUCGUAUGGAU_________GGUGAUGAUGGCGGAAG__AACAAGUGUUUUGCCAGCCCAAGCCCAAAUGUGAUUCAAUGCGCUGGGAAUCCAAAA ...(((((((((....((((((((..........(((.....((((((((...........))))))))......)))..........))))))))....))))))))). (-23.10 = -24.47 + 1.37)

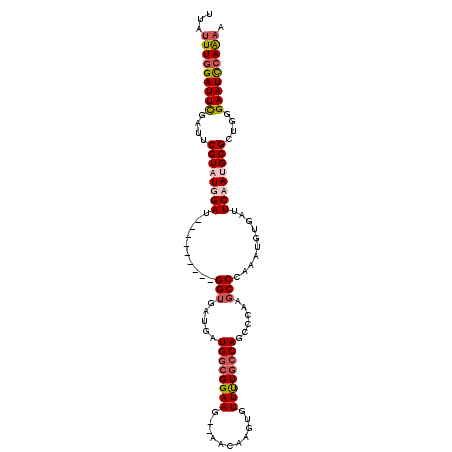
| Location | 17,549,575 – 17,549,667 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 90.05 |
| Mean single sequence MFE | -16.41 |
| Consensus MFE | -12.61 |
| Energy contribution | -12.42 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17549575 92 + 22224390 GUCUGAGCGGAAUGUAAACACAUUUUC-----------------AAAAGCCAAACAAAAGCCAAAUGAAAACAAUUCACACCCGAGGAAUUCGACUAAUAAUCCGCAAA ......(((((.(((.......(((((-----------------(...((.........))....)))))).(((((.(......))))))......))).)))))... ( -12.90) >DroSec_CAF1 1433 92 + 1 GUCUGAGCGGAAUGUAAACACAUUUUC-----------------AAAAGCCAAACAAAAGCCAAAUGAAAACAAUUCACACCCGAGGAAUUCGACUAACAAUCCGCAAA ......(((((.(((.......(((((-----------------(...((.........))....)))))).(((((.(......))))))......))).)))))... ( -14.80) >DroSim_CAF1 586 92 + 1 GUCUGAGCGGAAUGUAAACACAUUUUC-----------------AAAAGCCAAACAAAAGCCAAAUGAAAACAAUUCACACCCGAGGAAUUCGACUAACAAUCCGCAAA ......(((((.(((.......(((((-----------------(...((.........))....)))))).(((((.(......))))))......))).)))))... ( -14.80) >DroEre_CAF1 6104 107 + 1 GUCUGAGCGGAAUGUAAACACAUUUUCACCGGGCCUGCGUAUGCAAAAGCCAAACAAAAGCCAAAUGAAAACAAUUCACACCCGAGGAAUUCGACUAACAAUCCGCA-- ......(((((.(((.........(((..((((..(((....)))....................((((.....))))..))))..)))........))).))))).-- ( -23.13) >consensus GUCUGAGCGGAAUGUAAACACAUUUUC_________________AAAAGCCAAACAAAAGCCAAAUGAAAACAAUUCACACCCGAGGAAUUCGACUAACAAUCCGCAAA ......(((((.(((..................................................((((.....))))....((((...))))....))).)))))... (-12.61 = -12.42 + -0.19)



| Location | 17,549,575 – 17,549,667 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 90.05 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -19.35 |
| Energy contribution | -19.35 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17549575 92 - 22224390 UUUGCGGAUUAUUAGUCGAAUUCCUCGGGUGUGAAUUGUUUUCAUUUGGCUUUUGUUUGGCUUUU-----------------GAAAAUGUGUUUACAUUCCGCUCAGAC ...(((((.......((((.....))))((((((((((((((((...((((.......))))..)-----------------))))))).)))))))))))))...... ( -25.60) >DroSec_CAF1 1433 92 - 1 UUUGCGGAUUGUUAGUCGAAUUCCUCGGGUGUGAAUUGUUUUCAUUUGGCUUUUGUUUGGCUUUU-----------------GAAAAUGUGUUUACAUUCCGCUCAGAC ...(((((.......((((.....))))((((((((((((((((...((((.......))))..)-----------------))))))).)))))))))))))...... ( -25.60) >DroSim_CAF1 586 92 - 1 UUUGCGGAUUGUUAGUCGAAUUCCUCGGGUGUGAAUUGUUUUCAUUUGGCUUUUGUUUGGCUUUU-----------------GAAAAUGUGUUUACAUUCCGCUCAGAC ...(((((.......((((.....))))((((((((((((((((...((((.......))))..)-----------------))))))).)))))))))))))...... ( -25.60) >DroEre_CAF1 6104 107 - 1 --UGCGGAUUGUUAGUCGAAUUCCUCGGGUGUGAAUUGUUUUCAUUUGGCUUUUGUUUGGCUUUUGCAUACGCAGGCCCGGUGAAAAUGUGUUUACAUUCCGCUCAGAC --.(((((.......((((.....))))(((((((((((((((((..(((((.(((...((....))..))).)))))..))))))))).)))))))))))))...... ( -36.40) >consensus UUUGCGGAUUGUUAGUCGAAUUCCUCGGGUGUGAAUUGUUUUCAUUUGGCUUUUGUUUGGCUUUU_________________GAAAAUGUGUUUACAUUCCGCUCAGAC ...(((((.......((((.....))))(((((((((((((((....((((.......))))....................))))))).)))))))))))))...... (-19.35 = -19.35 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:55 2006