| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,543,924 – 17,544,164 |
| Length | 240 |
| Max. P | 0.968997 |

| Location | 17,543,924 – 17,544,044 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -22.36 |
| Energy contribution | -22.43 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17543924 120 + 22224390 CUUAGCCAAGACGCGAACCCAUACAGGUGCGUGUUACCCAGACAGCAACCGACACAGAAGUACCGCAUUUCGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGC .........((((((.(((......))).)))))).........((..........(((((.....)))))((((((...........))))))..((((((((.....)))))))).)) ( -31.60) >DroSec_CAF1 36041 120 + 1 UUUAGCGAAGAUGCGAACCCACACAGAUGCGUUUAUCCCAGACAUCAACCGACACAGAAGUGCCGCAUUUUGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGC ....(((((((((...(((.(((.((((((((((.....)))).......(.(((....))).)))))))))).)))..)))))))..........((((((((.....)))))))).)) ( -33.30) >DroSim_CAF1 25554 120 + 1 UUUAGCGAAGACGCGAACCCACACAGAUGCGUUUAUCCCAGACAUCAACCGACACAGAAGUGCCGCAUUUUGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGC ....(((....)))...........(((((((((.....)))).......(.(((....))).))))))((((((((...........))))))))((((((((.....))))))))... ( -31.20) >DroYak_CAF1 32861 117 + 1 GUUAGCUAAGACACGAACCGAAACAAGUGCACCUUCCCCAGACAGCAACCGAUAUAGAAAUAGCACAUUUCGUUGGUA---UUUUCAUACCAACAAUUGGCGCGAAAGCCGUGCCAAAGC ....(((............((((...((((..((.....)).............((....)))))).))))(((((((---(....))))))))..((((((((.....))))))))))) ( -28.50) >consensus UUUAGCGAAGACGCGAACCCACACAGAUGCGUUUAUCCCAGACAGCAACCGACACAGAAGUACCGCAUUUCGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGC ....((..................((((((((((.....)))).......(.(((....))).))))))).((((((...........))))))..((((((((.....)))))))).)) (-22.36 = -22.43 + 0.06)


| Location | 17,543,924 – 17,544,044 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -41.68 |
| Consensus MFE | -30.80 |
| Energy contribution | -30.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17543924 120 - 22224390 GCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACGAAAUGCGGUACUUCUGUGUCGGUUGCUGUCUGGGUAACACGCACCUGUAUGGGUUCGCGUCUUGGCUAAG ((.(((((((((...(((((....((((((((((.......))))))))))..))))).....)))))))))..))....(((.(((.((((((((....))))..)))).))).))).. ( -40.70) >DroSec_CAF1 36041 120 - 1 GCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACAAAAUGCGGCACUUCUGUGUCGGUUGAUGUCUGGGAUAAACGCAUCUGUGUGGGUUCGCAUCUUCGCUAAA ((.(((((.((.....)).)))))((((((((((.......))))))))))...))(((((....)))))(((.(((((...((((..((((....))))..)))))))))...)))... ( -41.60) >DroSim_CAF1 25554 120 - 1 GCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACAAAAUGCGGCACUUCUGUGUCGGUUGAUGUCUGGGAUAAACGCAUCUGUGUGGGUUCGCGUCUUCGCUAAA ((.(((((.((.....)).)))))((((((((((.......))))))))))...))(((((....)))))(((.(((((...((((..((((....))))..)))))))))...)))... ( -41.20) >DroYak_CAF1 32861 117 - 1 GCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUAUGAAAA---UACCAACGAAAUGUGCUAUUUCUAUAUCGGUUGCUGUCUGGGGAAGGUGCACUUGUUUCGGUUCGUGUCUUAGCUAAC (((..((((((((....))(((....((((((((....)---)))))))(((((((((..(((((....(((...)))....)))))...))))..)))))))).))))))..))).... ( -43.20) >consensus GCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACAAAAUGCGGCACUUCUGUGUCGGUUGAUGUCUGGGAAAAACGCACCUGUGUGGGUUCGCGUCUUCGCUAAA ...((((((((((..((..((...((((((((((.......))))))))))...))(((((....)))))))..))))).((.((..(((((....)))))...)).)).....))))). (-30.80 = -30.80 + 0.00)


| Location | 17,543,964 – 17,544,084 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -35.85 |
| Consensus MFE | -30.02 |
| Energy contribution | -29.65 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17543964 120 + 22224390 GACAGCAACCGACACAGAAGUACCGCAUUUCGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCAGAGAUGCGAUAAAGAAGAGGAGUGCAGAGAGGCGGCAGU ....((..((.((......))...(((((((((((((...........))))))..((((((((.....)))))))).(((....)))...........))))))).....))..))... ( -36.50) >DroSec_CAF1 36081 120 + 1 GACAUCAACCGACACAGAAGUGCCGCAUUUUGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCAGAGAUGCGAUAAAGAAGAGGAGUGCAGAGAGGCGGCAGU ....................((((((.((((....(((((.(((((..........((((((((.....)))))))).(((....)))......)))))..))))).)))).)))))).. ( -36.90) >DroSim_CAF1 25594 120 + 1 GACAUCAACCGACACAGAAGUGCCGCAUUUUGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCAGAGAUGCGAUAAAGAAGAGGAGUGCAGAGAGGCGGCAGU ....................((((((.((((....(((((.(((((..........((((((((.....)))))))).(((....)))......)))))..))))).)))).)))))).. ( -36.90) >DroYak_CAF1 32901 112 + 1 GACAGCAACCGAUAUAGAAAUAGCACAUUUCGUUGGUA---UUUUCAUACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCGGAGAUGCGAUUAAGAAGAGGCGCGCAGA-----GGCAGU ....((..(((.....(((((.....)))))(((((((---(....))))))))..((((((((.....))))))))..)))...((((.((......))...))))..-----.))... ( -33.10) >consensus GACAGCAACCGACACAGAAGUACCGCAUUUCGUUGGUAUUAUUUUCACACCAACAAUUGGCGCGAAAGCCGUGCCAAAGCAGAGAUGCGAUAAAGAAGAGGAGUGCAGAGAGGCGGCAGU ........((..(((....))).((((((((((((((...........))))))..((((((((.....))))))))....))))))))..........))...((.........))... (-30.02 = -29.65 + -0.37)


| Location | 17,543,964 – 17,544,084 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -34.84 |
| Consensus MFE | -30.61 |
| Energy contribution | -29.68 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17543964 120 - 22224390 ACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACGAAAUGCGGUACUUCUGUGUCGGUUGCUGUC ((.((.(((.....((((..........((((((((.((....(((((.((.....)).)))))..((((((((.......)))))))))).)))))))).....)))).))).)).)). ( -35.66) >DroSec_CAF1 36081 120 - 1 ACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACAAAAUGCGGCACUUCUGUGUCGGUUGAUGUC .(.(((((......))................((((.......(((((.((.....)).)))))((((((((((.......)))))))))).))))(((((....)))))))).)..... ( -35.30) >DroSim_CAF1 25594 120 - 1 ACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACAAAAUGCGGCACUUCUGUGUCGGUUGAUGUC .(.(((((......))................((((.......(((((.((.....)).)))))((((((((((.......)))))))))).))))(((((....)))))))).)..... ( -35.30) >DroYak_CAF1 32901 112 - 1 ACUGCC-----UCUGCGCGCCUCUUCUUAAUCGCAUCUCCGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUAUGAAAA---UACCAACGAAAUGUGCUAUUUCUAUAUCGGUUGCUGUC ...((.-----...))(((((...........((......))..(((((((((......)))..((((((((((....)---)))))))))...))))))..........)).))).... ( -33.10) >consensus ACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAUAAUACCAACAAAAUGCGGCACUUCUGUGUCGGUUGAUGUC .(.(((..........................((((.......(((((.((.....)).)))))((((((((((.......)))))))))).))))(((((....)))))))).)..... (-30.61 = -29.68 + -0.94)

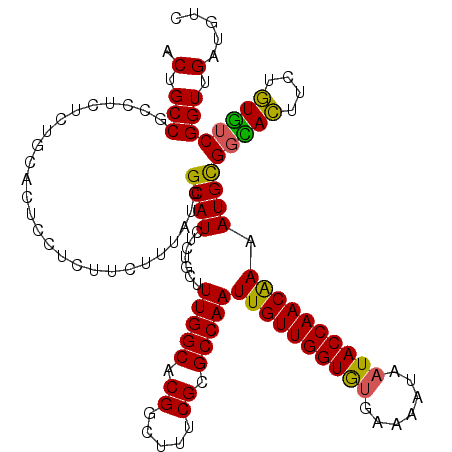

| Location | 17,544,004 – 17,544,124 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -34.05 |
| Energy contribution | -34.62 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17544004 120 - 22224390 CGCCGCGCUUGUGUGCGAUUCCACUGGUGUUUCUCCACAGACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAU .((((.(((.(((((((((......(((((((......)))).)))((......)).............)))))))....))...))).))))((((((((((.....)))))))))).. ( -37.60) >DroSec_CAF1 36121 120 - 1 CGCCGCGCUUGUGUGUGAUUCCACUGGUGUUUCUCCGCAGACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAU .((((.(((.(((((((((......((.......))(((((..(....)..))))).............)))))))....))...))).))))((((((((((.....)))))))))).. ( -37.80) >DroSim_CAF1 25634 120 - 1 CGCCGCGCUUGUGUGUGAUUCCACUGGUGUUUCUCCGCAGACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAU .((((.(((.(((((((((......((.......))(((((..(....)..))))).............)))))))....))...))).))))((((((((((.....)))))))))).. ( -37.80) >DroYak_CAF1 32939 114 - 1 CGCCGCGCUUGUGUGCGACUCCGCUGGUGUUUCUCCACAGACUGCC-----UCUGCGCGCCUCUUCUUAAUCGCAUCUCCGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUAUGAAAA- .((((.(((.((((((((.......(((((.......((((.....-----)))).))))).........)))))....)))...))).))))(((((..(((.....)))..))))).- ( -32.89) >consensus CGCCGCGCUUGUGUGCGAUUCCACUGGUGUUUCUCCACAGACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCUGCUUUGGCACGGCUUUCGCGCCAAUUGUUGGUGUGAAAAU .((((.(((.(((((((((......((.((......(((((..........))))))).))........)))))))....))...))).))))((((((((((.....)))))))))).. (-34.05 = -34.62 + 0.56)

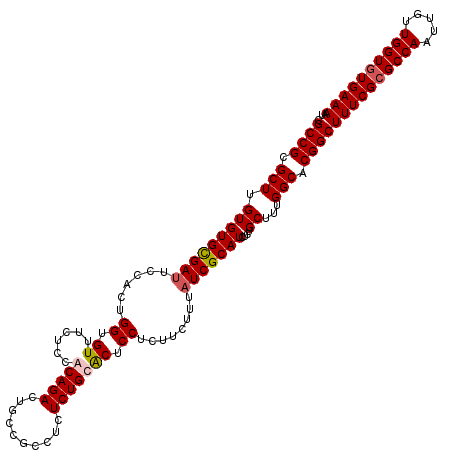

| Location | 17,544,044 – 17,544,164 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -27.97 |
| Energy contribution | -28.34 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17544044 120 - 22224390 AACCCGCGGUCUGGAUUUUCACACACGUCCGCUUUCGAAUCGCCGCGCUUGUGUGCGAUUCCACUGGUGUUUCUCCACAGACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCU .....((((((((((...((.((((((..(((............)))..)))))).)).)))....(((......)))))))))).((......))........................ ( -30.20) >DroSec_CAF1 36161 120 - 1 AACCCGCGGCCUGGAUUUUCACACACGUCCGCUUUCGAAUCGCCGCGCUUGUGUGUGAUUCCACUGGUGUUUCUCCGCAGACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCU .....((((((((((...(((((((((..(((............)))..))))))))).))))..)))........(((((..(....)..)))))...............)))...... ( -35.40) >DroSim_CAF1 25674 120 - 1 AACCCGCGGCCUGGAUUUUCACACACGUCCGCUUUCGAAUCGCCGCGCUUGUGUGUGAUUCCACUGGUGUUUCUCCGCAGACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCU .....((((((((((...(((((((((..(((............)))..))))))))).))))..)))........(((((..(....)..)))))...............)))...... ( -35.40) >DroYak_CAF1 32978 115 - 1 AACCCGCGGCCUGGAUUUUCACACACGUCCGCUUUCGAAUCGCCGCGCUUGUGUGCGACUCCGCUGGUGUUUCUCCACAGACUGCC-----UCUGCGCGCCUCUUCUUAAUCGCAUCUCC .....((((...(((...((.((((((..(((............)))..)))))).)).)))((.(((((((......)))).)))-----...))..............))))...... ( -27.10) >consensus AACCCGCGGCCUGGAUUUUCACACACGUCCGCUUUCGAAUCGCCGCGCUUGUGUGCGAUUCCACUGGUGUUUCUCCACAGACUGCCGCCUCUCUGCACUCCUCUUCUUUAUCGCAUCUCU .....(((((.((((...(((((((((..(((............)))..))))))))).))))(((.((......)))))...))))).....(((................)))..... (-27.97 = -28.34 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:50 2006