| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,542,060 – 17,542,169 |
| Length | 109 |
| Max. P | 0.738378 |

| Location | 17,542,060 – 17,542,169 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.59 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -20.31 |
| Energy contribution | -20.78 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
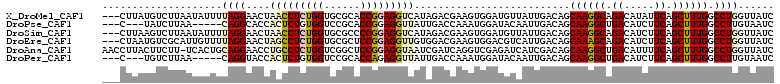
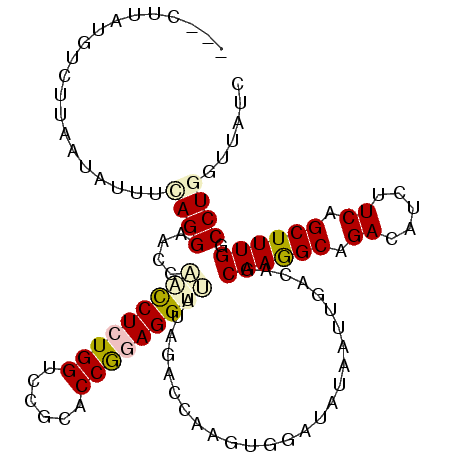
>X_DroMel_CAF1 17542060 109 - 22224390 ---CUUAUGUCUUAAUAUUUUAGGAACUAACCUCUGGUGCGCACCGGAGGUCAUAGACGAAGUGGAUGUUAUUGACAGCAAGGCAGACAUAUUCAGCUUUGGCCUGGUUAUC ---......((((((....))))))..(((((((((((....))))))(((((.((..((((((..((((.(((....)))))))..))).)))..)).))))).))))).. ( -31.40) >DroPse_CAF1 36401 101 - 1 ---C---UAUCUUAA-----CAGGCACCACUCUGUGGUCCGCACCGGAGGUUAUUGACCAAAUGGAUACAAUUGACAGCAAGGCUGACAUCUUCAGCUUUGGCCUUGUAAUC ---.---.......(-----(((((.(((.....(((((...(((...)))....)))))..))).............(((((((((.....)))))))))))).))).... ( -28.10) >DroSim_CAF1 23660 109 - 1 ---CUUAAGUCUUAAUAUUUUAGGAACUAACCUCUGGUGCGCCCCGGAGGUCAUAGACGAAGUGGAUGUUAUUGACAGCAAGGCAGACAUCUUCAGCUUUGGCCUGGUUAUC ---......((((((....))))))..((((((((((......)))))(((((.((..((((((..((((.(((....)))))))..)).))))..)).))))).))))).. ( -31.50) >DroEre_CAF1 29835 109 - 1 ---CUAAUGUCGCAUUGUUUUAGGAACUAGCCUCUGGUGCGCUCCGGAGGUUGUGGACGAAGUGGACGUCAUUGACAGCAAAGCAGACAUCUUCAGCUUUGGCCUGGUUAUC ---...(((((((.(((((((((....((((((((((......))))))))))..((((.......)))).)))).))))).)).)))))..(((((....).))))..... ( -31.40) >DroAna_CAF1 24788 111 - 1 AACCUUACUUCUU-UCACUGCAGGAACCUGCCUCUGGUCGGCUCCGGAGGUAAUCGAUCAGGUCGAGAUCAUCGACAGCAAGGCUGACAUUUUCAGCUUUGGCCUGGUUAUC ....((((.....-.....((((....))))((((((......))))))))))..(((((((((((.....)))))..(((((((((.....)))))))))..))))))... ( -40.20) >DroPer_CAF1 35672 101 - 1 ---C---UGUCUUAA-----CAGGUACCACUCUGUGGUCCGCACCAGAGGUUAUUGACCAAAUGGAUACAAUUGACAGCAAGGCUGACAUCUUCAGCUUUGGCCUUGUAAUC ---(---((((....-----..((.(((((...)))))))...(((..((((...))))...)))........)))))(((((((((.....)))))))))........... ( -31.20) >consensus ___CUUAUGUCUUAAUAUUUCAGGAACCAACCUCUGGUCCGCACCGGAGGUUAUAGACCAAGUGGAUAUAAUUGACAGCAAGGCAGACAUCUUCAGCUUUGGCCUGGUUAUC ....................((((....(((((((((......)))))))))..........................((((((.((.....)).)))))).))))...... (-20.31 = -20.78 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:45 2006