
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,535,981 – 17,536,095 |
| Length | 114 |
| Max. P | 0.815063 |

| Location | 17,535,981 – 17,536,095 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.18 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -15.34 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17535981 114 + 22224390 ACGCGAUUUUAAGCUCUCACUACUAAACCAUUCAUAAGAUUAAGAAGCAUCUGUUGGAUAGGAUUCUUUACAAUCACGGAUACAACUUCUUAAGAAUCGUUUCGUAUUAUUAUU ..((........))......(((.((((.((((.(((((...((..(.((((((..(((.(.........).))))))))).)..))))))).)))).)))).)))........ ( -17.60) >DroSec_CAF1 28283 106 + 1 AUGGGUUUUUAUGCUCUCACUACUA-ACCAUUCUUAAGAUUUAGAAGCAUCU---GGCUAGGAUUCUUAACAACCACGGAUACAACUUCUUAAGAAUGGUUUCGUAUUAU---- ..((((......))))....(((.(-(((((((((((((...((..(.((((---(....((...........)).))))).)..))))))))))))))))..)))....---- ( -25.40) >DroSim_CAF1 17719 106 + 1 AUGGGUUUUUAUGCUCUCACUACUA-ACCAUUCUUAAGAUUUAGAAGCAUCU---GGCUAGGAUUCUUAACAAACACGGAUACAACUUCUUAAGAAUGGUUUCGUAUUAU---- ..((((......))))....(((.(-(((((((((((((...((..(.((((---(.....(........).....))))).)..))))))))))))))))..)))....---- ( -25.00) >DroEre_CAF1 23814 99 + 1 -------AUGGUGCUCUCAGUACCA-GCCAUACUUAAGAUUUAGAAGCAUCU---GGCUCGGAUUCUUUACAAUCAUUGGUACAACUUCUUAAGAAUGGUUUCGUAUAAU---- -------.((((((.....))))))-(((((.(((((((....((((.((((---(...))))).)))).((.....))........))))))).)))))..........---- ( -24.60) >consensus AUGGGUUUUUAUGCUCUCACUACUA_ACCAUUCUUAAGAUUUAGAAGCAUCU___GGCUAGGAUUCUUAACAAUCACGGAUACAACUUCUUAAGAAUGGUUUCGUAUUAU____ ....................(((...(((((((((((((.....(((.((((........)))).)))...................)))))))))))))...)))........ (-15.34 = -15.90 + 0.56)

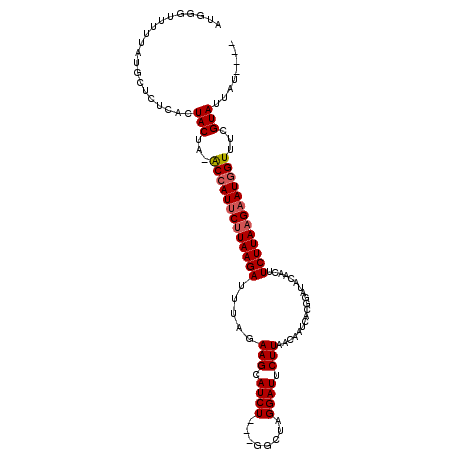
| Location | 17,535,981 – 17,536,095 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.18 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -16.91 |
| Energy contribution | -18.41 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17535981 114 - 22224390 AAUAAUAAUACGAAACGAUUCUUAAGAAGUUGUAUCCGUGAUUGUAAAGAAUCCUAUCCAACAGAUGCUUCUUAAUCUUAUGAAUGGUUUAGUAGUGAGAGCUUAAAAUCGCGU ........(((.((((.((((.(((((((..(((((.((((((......)))).......)).)))))..))...))))).)))).)))).)))((((..........)))).. ( -21.51) >DroSec_CAF1 28283 106 - 1 ----AUAAUACGAAACCAUUCUUAAGAAGUUGUAUCCGUGGUUGUUAAGAAUCCUAGCC---AGAUGCUUCUAAAUCUUAAGAAUGGU-UAGUAGUGAGAGCAUAAAAACCCAU ----....(((..((((((((((((((((..(((((..((((((..........)))))---))))))..))...)))))))))))))-).)))((....))............ ( -31.30) >DroSim_CAF1 17719 106 - 1 ----AUAAUACGAAACCAUUCUUAAGAAGUUGUAUCCGUGUUUGUUAAGAAUCCUAGCC---AGAUGCUUCUAAAUCUUAAGAAUGGU-UAGUAGUGAGAGCAUAAAAACCCAU ----....(((..((((((((((((((((..(((((.(.(((.............))))---.)))))..))...)))))))))))))-).)))((....))............ ( -27.52) >DroEre_CAF1 23814 99 - 1 ----AUUAUACGAAACCAUUCUUAAGAAGUUGUACCAAUGAUUGUAAAGAAUCCGAGCC---AGAUGCUUCUAAAUCUUAAGUAUGGC-UGGUACUGAGAGCACCAU------- ----...........((((.(((((((............((((......)))).((((.---....)))).....))))))).)))).-((((.(.....).)))).------- ( -17.20) >consensus ____AUAAUACGAAACCAUUCUUAAGAAGUUGUAUCCGUGAUUGUAAAGAAUCCUAGCC___AGAUGCUUCUAAAUCUUAAGAAUGGU_UAGUAGUGAGAGCAUAAAAACCCAU ........(((...(((((((((((((((..(((((...........................)))))..))...)))))))))))))...)))((....))............ (-16.91 = -18.41 + 1.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:44 2006