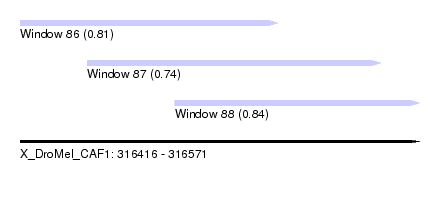
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 316,416 – 316,571 |
| Length | 155 |
| Max. P | 0.844556 |

| Location | 316,416 – 316,516 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -28.50 |
| Energy contribution | -29.10 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 316416 100 + 22224390 GAA--------------UGAGGACGUGGCAUCCUGGAUCAAAAACCGGUAGUCGGUCGUCGCAAGUUUCUUCUCCCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACC (((--------------.((((((.((((.....((((....((((((..(..((..(.(....)...)..))..)..))))))))))...)).)).)))))))))........ ( -28.80) >DroSec_CAF1 59388 100 + 1 GAA--------------UGAGGACUUGGCAUCCUGGAUCGAAAACCGGUAGACGGUCGUCGCAAGUUUCUUCUCCCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACC (((--------------.(((((((((((.....((((....((((((((((.((....(....)..)).)))....)))))))))))...)).))))))))))))........ ( -32.00) >DroSim_CAF1 80023 100 + 1 GAA--------------UGAGGACUUGGCAUCCUGGAUCGAAAACCGGUAGACGGUCGUCGCAAGUUUCUUCUCCCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACC (((--------------.(((((((((((.....((((....((((((((((.((....(....)..)).)))....)))))))))))...)).))))))))))))........ ( -32.00) >DroEre_CAF1 60064 100 + 1 GAA--------------UGAGGACUUGGCAUCCUGGAUCGAAAACCGGUAGUCGUUCGUCGCAAGUUUCUUCUCCCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACC (((--------------.(((((((((((.....((((....((((((..(........(....)..........)..))))))))))...)).))))))))))))........ ( -29.97) >DroYak_CAF1 80781 114 + 1 GAAUGAGGAAUAAGGAAUGAGGACUUGGCAUCCUGGAUCGCAAACCGGUAGUCGUUCGUCGCAAGUUUCUGCUCCCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACC ......((..(((((((.(((((((((((.....((((....(((((((.(..((....(....).....))..)..)))))))))))...)).))))))))))))))))..)) ( -39.50) >consensus GAA______________UGAGGACUUGGCAUCCUGGAUCGAAAACCGGUAGUCGGUCGUCGCAAGUUUCUUCUCCCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACC ..................(((((((((((.....((((....((((((..(..((....(....)......))..)..))))))))))...)).)))))))))........... (-28.50 = -29.10 + 0.60)


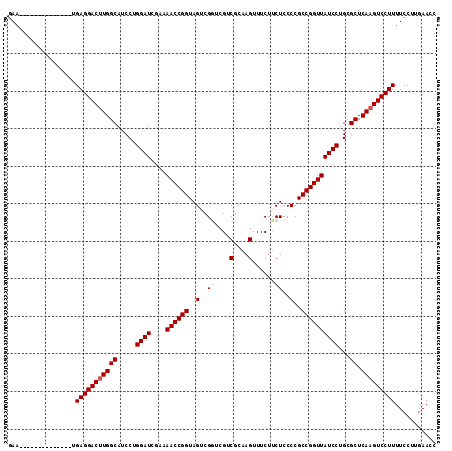
| Location | 316,442 – 316,556 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 97.89 |
| Mean single sequence MFE | -27.89 |
| Consensus MFE | -26.60 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 316442 114 + 22224390 AAAACCGGUAGUCGGUCGUCGCAAGUUUCUUCUCCCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACCUUUUAAGUGAACUCAAGUUUAUAAAUUGUGCAGCAAGUAC ..((((((..(..((..(.(....)...)..))..)..))))))...(((((((((((........))))).........((((((....)))))).....))))))....... ( -29.10) >DroSec_CAF1 59414 114 + 1 AAAACCGGUAGACGGUCGUCGCAAGUUUCUUCUCCCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACCUUUCAAGUGAACUCAAGUUUAUAAAUUGUGCAGCAAGUAC ..((((((((((.((....(....)..)).)))....)))))))...(((((((((((........))))).........((((((....)))))).....))))))....... ( -28.20) >DroSim_CAF1 80049 114 + 1 AAAACCGGUAGACGGUCGUCGCAAGUUUCUUCUCCCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACCUUUCAAGUGAACUCAAGUUUAUAAAUUGUGCAGCAAGUAC ..((((((((((.((....(....)..)).)))....)))))))...(((((((((((........))))).........((((((....)))))).....))))))....... ( -28.20) >DroEre_CAF1 60090 114 + 1 AAAACCGGUAGUCGUUCGUCGCAAGUUUCUUCUCCCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACCUUUCAAGUGAACUCAAGUUUAUAAAUUGUGCAGCAAGUAC ..((((((..(........(....)..........)..))))))...(((((((((((........))))).........((((((....)))))).....))))))....... ( -26.17) >DroYak_CAF1 80821 114 + 1 CAAACCGGUAGUCGUUCGUCGCAAGUUUCUGCUCCCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACCUUUCAAGUGAACUCAAGUUUAUAAAUUGUGCAGCAAGUAC ..(((((((.(..((....(....).....))..)..)))))))...(((((((((((........))))).........((((((....)))))).....))))))....... ( -27.80) >consensus AAAACCGGUAGUCGGUCGUCGCAAGUUUCUUCUCCCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACCUUUCAAGUGAACUCAAGUUUAUAAAUUGUGCAGCAAGUAC ..((((((..(..((....(....)......))..)..))))))...(((((((((((........))))).........((((((....)))))).....))))))....... (-26.60 = -27.00 + 0.40)

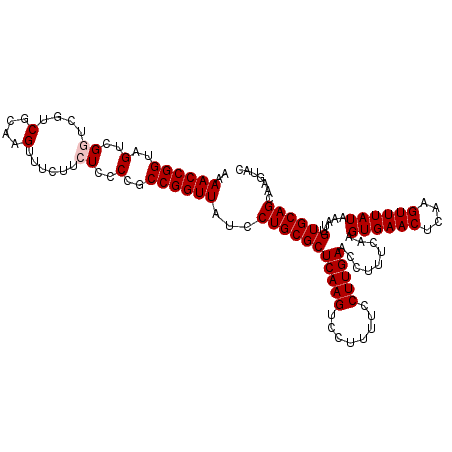
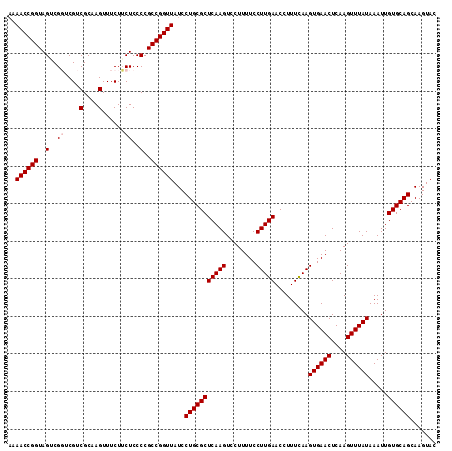
| Location | 316,476 – 316,571 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 93.18 |
| Mean single sequence MFE | -18.32 |
| Consensus MFE | -16.90 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 316476 95 + 22224390 CCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACCUUUUAAGUGAACUCAAGUUUAUAAAUUGUGCAGCAAGUACAACACAC--ACACA--------UAU ..(((.((....)).))).(((((........))))).........((((((....))))))...((((((.....)))))).....--.....--------... ( -16.90) >DroSec_CAF1 59448 97 + 1 CCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACCUUUCAAGUGAACUCAAGUUUAUAAAUUGUGCAGCAAGUACAACACACACACACA--------UAU ..(((.((....)).))).(((((........))))).........((((((....))))))...((((((.....))))))............--------... ( -16.90) >DroSim_CAF1 80083 97 + 1 CCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACCUUUCAAGUGAACUCAAGUUUAUAAAUUGUGCAGCAAGUACAACACACACACACA--------UAU ..(((.((....)).))).(((((........))))).........((((((....))))))...((((((.....))))))............--------... ( -16.90) >DroEre_CAF1 60124 105 + 1 CCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACCUUUCAAGUGAACUCAAGUUUAUAAAUUGUGCAGCAAGUACAACACACACGCACACAUACGUGUAU ..(((.((....)).))).(((((........))))).........((((((....))))))...((((((.....))))))...(((((........))))).. ( -21.80) >DroYak_CAF1 80855 105 + 1 CCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACCUUUCAAGUGAACUCAAGUUUAUAAAUUGUGCAGCAAGUACAACACACACAUACAUAUACUUGUAU ..(((.((....)).))).(((((........))))).........((((((....))))))...((((((.....)))))).......(((((......))))) ( -19.10) >consensus CCCGCCGGUUAUCCUGCGCUCAAGUCCUUUUCCUUGAACCUUUCAAGUGAACUCAAGUUUAUAAAUUGUGCAGCAAGUACAACACACACACACA________UAU ..(((.((....)).))).(((((........))))).........((((((....))))))...((((((.....))))))....................... (-16.90 = -16.90 + 0.00)

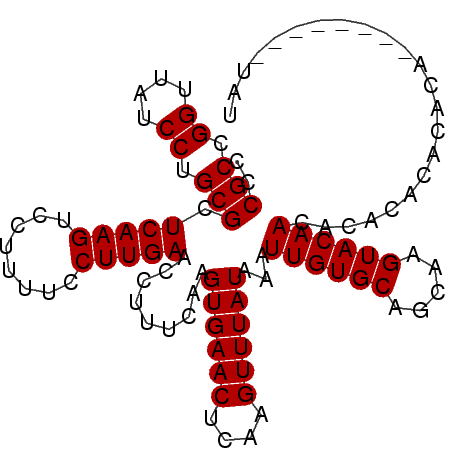
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:03 2006