
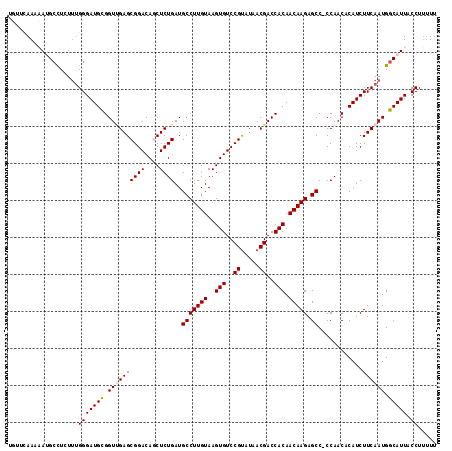
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,516,107 – 17,516,223 |
| Length | 116 |
| Max. P | 0.881910 |

| Location | 17,516,107 – 17,516,223 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 91.16 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -24.52 |
| Energy contribution | -24.68 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17516107 116 + 22224390 UGUUCAAAAAUGCCUGUUUGGGAUGCGGUUGAGCGGACAGCUCUGAUGCCUUGUAAGUGUCCGUAUAACGACCACAACAAGAGCCCCCAACACAUCUUCAAUGGCAUUACCUUUUU ........((((((.(((..(((((.(...((((.....))))....(((((((..(((..((.....))..))).))))).))......).)))))..)))))))))........ ( -32.00) >DroSec_CAF1 7016 115 + 1 UUUUCAAAAAUGCCUCUUUGGGAUGCGGUUGAGCGGACAGCUCUGAUGCCUUGUAAGUGUCCGUAUAACGACCACAACAAGAGCC-CCAACACAUCUUCAAUGGCAUUACCUUUUU ........((((((...((((((((.(...((((.....))))((..(((((((..(((..((.....))..))).))))).)).-.)).).)))).)))).))))))........ ( -32.40) >DroSim_CAF1 5332 114 + 1 UUUUCAAAAAUGCCUCUUUGGGAUGCGGUUGAGCGGACAGCUCUGAUGCCUUGUAAGUGUCCGUAUAACGACCACAACAAGAGCC-CCAACACAUCUUCAAUGGCAUUACCUU-UU ........((((((...((((((((.(...((((.....))))((..(((((((..(((..((.....))..))).))))).)).-.)).).)))).)))).)))))).....-.. ( -32.40) >DroEre_CAF1 5019 115 + 1 AGUCGAAGUAAGCUUGUUUGGGAUGUGGUGGAGCGGACAUCUCUGAUGCCUUGUAAGUGUCCGUAUAACGACCACAACAAGAGCC-CCAACACAUCUUCAAUGGCAUUACCUUUUU .......(((((((...((((((((((.(((.((((.((((...))))))((((..(((..((.....))..))).))))..)).-))).)))))).)))).))).))))...... ( -33.60) >DroYak_CAF1 5306 114 + 1 UGUUUAA-UAAGGUUUUCAGGGAUGUGGUCGAGCGGACAACUCUGAUGCCUUGUAAGUGUCCGCAUAACGACCACAACAAGAGCC-CCAACACAUCUUCAAUGGCAUUACCUCUUU ((((...-.(((((.....(((.((((((((.(((((((.((...((.....)).)))))))))....)))))))).......))-)......)))))....)))).......... ( -29.20) >consensus UGUUCAAAAAUGCCUCUUUGGGAUGCGGUUGAGCGGACAGCUCUGAUGCCUUGUAAGUGUCCGUAUAACGACCACAACAAGAGCC_CCAACACAUCUUCAAUGGCAUUACCUUUUU ...................(((((((.((((((((((....))))..(((((((..(((..((.....))..))).))))).))............)))))).))))).))..... (-24.52 = -24.68 + 0.16)


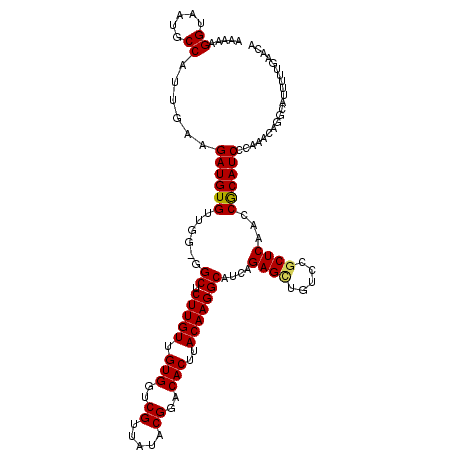
| Location | 17,516,107 – 17,516,223 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 91.16 |
| Mean single sequence MFE | -36.82 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.24 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17516107 116 - 22224390 AAAAAGGUAAUGCCAUUGAAGAUGUGUUGGGGGCUCUUGUUGUGGUCGUUAUACGGACACUUACAAGGCAUCAGAGCUGUCCGCUCAACCGCAUCCCAAACAGGCAUUUUUGAACA .....((.....)).(..(((((((.((((((((.(((((.(((..((.....))..)))..)))))))....((((.....)))).......))))))....)))))))..)... ( -37.30) >DroSec_CAF1 7016 115 - 1 AAAAAGGUAAUGCCAUUGAAGAUGUGUUGG-GGCUCUUGUUGUGGUCGUUAUACGGACACUUACAAGGCAUCAGAGCUGUCCGCUCAACCGCAUCCCAAAGAGGCAUUUUUGAAAA .....((.....)).(..(((((((.((((-(((.(((((.(((..((.....))..)))..)))))))....((((.....))))........)))))....)))))))..)... ( -36.60) >DroSim_CAF1 5332 114 - 1 AA-AAGGUAAUGCCAUUGAAGAUGUGUUGG-GGCUCUUGUUGUGGUCGUUAUACGGACACUUACAAGGCAUCAGAGCUGUCCGCUCAACCGCAUCCCAAAGAGGCAUUUUUGAAAA ..-..((.....)).(..(((((((.((((-(((.(((((.(((..((.....))..)))..)))))))....((((.....))))........)))))....)))))))..)... ( -36.60) >DroEre_CAF1 5019 115 - 1 AAAAAGGUAAUGCCAUUGAAGAUGUGUUGG-GGCUCUUGUUGUGGUCGUUAUACGGACACUUACAAGGCAUCAGAGAUGUCCGCUCCACCACAUCCCAAACAAGCUUACUUCGACU ....((((((.((..(((..((((((.(((-(((.(((((.(((..((.....))..)))..)))))(((((...)))))..)))))).)))))).)))....))))))))..... ( -37.40) >DroYak_CAF1 5306 114 - 1 AAAGAGGUAAUGCCAUUGAAGAUGUGUUGG-GGCUCUUGUUGUGGUCGUUAUGCGGACACUUACAAGGCAUCAGAGUUGUCCGCUCGACCACAUCCCUGAAAACCUUA-UUAAACA ...(((((.....(((.......)))(..(-((.......((((((((....((((((((((...........))).))))))).)))))))).)))..)..))))).-....... ( -36.20) >consensus AAAAAGGUAAUGCCAUUGAAGAUGUGUUGG_GGCUCUUGUUGUGGUCGUUAUACGGACACUUACAAGGCAUCAGAGCUGUCCGCUCAACCGCAUCCCAAACAGGCAUUUUUGAACA .....((.....))......((((((......((.(((((.(((..((.....))..)))..)))))))....((((.....))))...))))))..................... (-27.40 = -27.24 + -0.16)

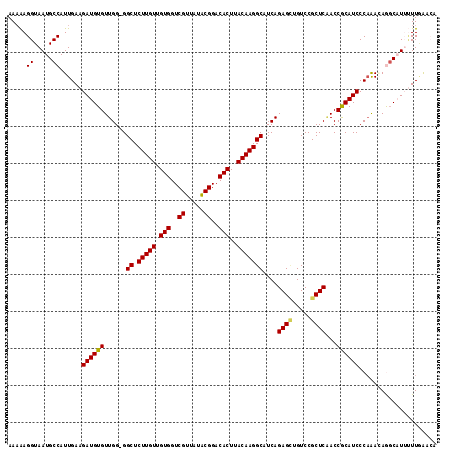
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:39 2006