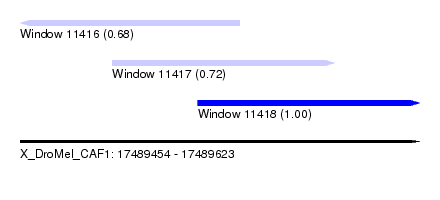
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,489,454 – 17,489,623 |
| Length | 169 |
| Max. P | 0.998636 |

| Location | 17,489,454 – 17,489,547 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 70.39 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -14.12 |
| Energy contribution | -13.38 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17489454 93 - 22224390 UUUGUGU----------------------AACAUGUGUUAAGCAAAAGCAAAAGCGAGAAAAUGUUGCUUAGCUAUCUCGCUCUAAUUCGCUAGAGCGAUUGAA-UCUUAUUUGGA (((((.(----------------------(((....)))).)))))(((..((((((.......)))))).)))...((((((((......)))))))).....-........... ( -23.20) >DroSec_CAF1 2632 87 - 1 AGUUUA-----------------------AAUAUGAGU------AAAGCAAAAGGGGGAAAUUGCCGCUUAGCUAUCUCCCUCUAAUUCGCUAGCGCGAUUGAUUUCUUAUUUGGA ..((((-----------------------((((.(((.------....(((.(((((((.((.((......)).)).)))))))...((((....)))))))..))).)))))))) ( -21.80) >DroSim_CAF1 2843 86 - 1 AGUUUA-----------------------AAUAAGCUU------AAAGCAAAAGGGGGAAAUUGCCGCUUAGCUAUCUCCCUCUAAUUCGCUAGCGCGAUUGAA-UCUUAUUUGGA ..((((-----------------------((((((.((------((......(((((((.((.((......)).)).)))))))...((((....)))))))).-.)))))))))) ( -22.70) >DroEre_CAF1 2642 109 - 1 UUUUUAAAACAUGUGUACAUUUUGACUGAAAUAUGCAU------AAAGCAGAAAGGGGGAAAUGCCGCUUAGCUGUCUCUCUCUAAUUCGCAAAAGCCAACGAA-UCAUAUUUGGA ...........((((((.(((((....))))).)))))------)..((.((((((((((...((......))..)))))))....))))).....((((....-......)))). ( -19.60) >consensus AGUUUA_______________________AAUAUGCGU______AAAGCAAAAGGGGGAAAAUGCCGCUUAGCUAUCUCCCUCUAAUUCGCUAGAGCGAUUGAA_UCUUAUUUGGA ................................................(((((((((((....((......))..))))))).....((((....))))...........)))).. (-14.12 = -13.38 + -0.75)

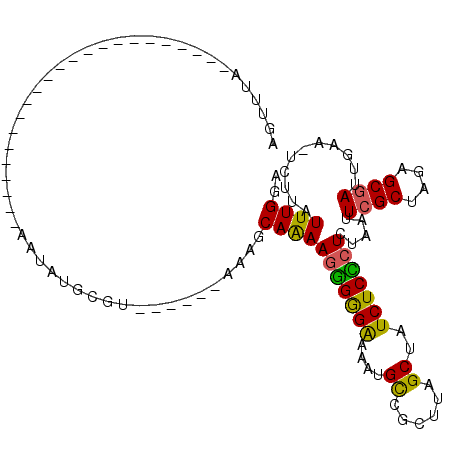
| Location | 17,489,493 – 17,489,587 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.07 |
| Mean single sequence MFE | -16.23 |
| Consensus MFE | -7.68 |
| Energy contribution | -8.87 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17489493 94 + 22224390 AUAGCUAAGCAACAUUUUCUCGCUUUUGCUUUUGCUUAACACAUGUU----------------------ACACAAAAAUAAGCAGAACUCCAAAUGAAUGGUACUCCAAACAAAAA ..(((.((((...........))))..)))((((((((.....((..----------------------...))....))))))))............(((....)))........ ( -12.80) >DroSec_CAF1 2672 87 + 1 AUAGCUAAGCGGCAAUUUCCCCCUUUUGCUUU------ACUCAUAUU-----------------------UAAACUAAUAAGCAGAACUACAAAUGAAUGGUUCUGCAAAUAAUAA ..(((.(((.((........)))))..)))..------.........-----------------------...........(((((((((........)))))))))......... ( -16.80) >DroSim_CAF1 2882 87 + 1 AUAGCUAAGCGGCAAUUUCCCCCUUUUGCUUU------AAGCUUAUU-----------------------UAAACUAAUAAGCAGAACUACAAAUGAAUGGUUCUGCAAAUAAUAA .(((.((((((((((..........)))))..------..))))).)-----------------------)).........(((((((((........)))))))))......... ( -20.10) >DroEre_CAF1 2681 110 + 1 ACAGCUAAGCGGCAUUUCCCCCUUUCUGCUUU------AUGCAUAUUUCAGUCAAAAUGUACACAUGUUUUAAAAAGAUAAGCUGAACUAUAAAUGCAUGUUUAUACAAACAGUAC .....((((((((((((..........((...------..))(((.((((((.....((....))(((((.....))))).)))))).))))))))).))))))............ ( -15.20) >consensus AUAGCUAAGCGGCAAUUUCCCCCUUUUGCUUU______ACGCAUAUU_______________________UAAAAAAAUAAGCAGAACUACAAAUGAAUGGUUCUGCAAACAAUAA ......((((((.............))))))..................................................(((((((((........)))))))))......... ( -7.68 = -8.87 + 1.19)



| Location | 17,489,529 – 17,489,623 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.82 |
| Mean single sequence MFE | -14.15 |
| Consensus MFE | -7.05 |
| Energy contribution | -9.30 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.50 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17489529 94 + 22224390 UAACACAUGUU----------------------ACACAAAAAUAAGCAGAACUCCAAAUGAAUGGUACUCCAAACAAAAAAGAUCAAUCAACAAAAUAUGAUCAAAAAACAUUUCU ((((....)))----------------------).....................(((((..(((....))).........(((((............)))))......))))).. ( -7.90) >DroSec_CAF1 2704 91 + 1 --ACUCAUAUU-----------------------UAAACUAAUAAGCAGAACUACAAAUGAAUGGUUCUGCAAAUAAUAAAGAUCAAUCAACAAGAUAUGAUCAAAAAACAUUUCU --.........-----------------------...........(((((((((........)))))))))..........((((((((.....))).)))))............. ( -19.00) >DroSim_CAF1 2914 91 + 1 --AAGCUUAUU-----------------------UAAACUAAUAAGCAGAACUACAAAUGAAUGGUUCUGCAAAUAAUAAAGAUCAAACAACAAGAUAUGAUCAAAAAACAUUUCU --..(((((((-----------------------......)))))))(((((((........)))))))............(((((............)))))............. ( -16.20) >DroEre_CAF1 2713 113 + 1 --AUGCAUAUUUCAGUCAAAAUGUACACAUGUUUUAAAAAGAUAAGCUGAACUAUAAAUGCAUGUUUAUACAAACAGUACAAAUCAAUUACAAAGGUGUGAUCAUAUAACA-AUCC --.(((((.((......)).)))))(((((.(((......(((..((((...(((((((....)))))))....))))....)))......))).)))))...........-.... ( -13.50) >consensus __ACGCAUAUU_______________________UAAAAAAAUAAGCAGAACUACAAAUGAAUGGUUCUGCAAACAAUAAAGAUCAAUCAACAAGAUAUGAUCAAAAAACAUUUCU .............................................(((((((((........)))))))))..........((((((((.....))).)))))............. ( -7.05 = -9.30 + 2.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:28 2006