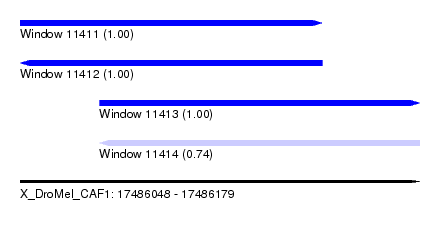
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,486,048 – 17,486,179 |
| Length | 131 |
| Max. P | 0.999966 |

| Location | 17,486,048 – 17,486,147 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.38 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.98 |
| SVM RNA-class probability | 0.999966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17486048 99 + 22224390 UCAAGUUUAAUCCGCCUCGGCGACACUUUUUCUUGAAGCCCGUGCUUCUUGGCUUUUCCAGCCAGUCACUUGACAAAAUACUAAGCAAUUGUAUUUGUU ((((((.......((....))(((..........(((((....))))).(((((.....))))))))))))))..((((((.((....))))))))... ( -25.80) >DroSec_CAF1 3937 99 + 1 UCAAGUUUAAUCCGCCUCGGCGACACUUUUUCUUGAAGCCCGUGCUUCUUGGCUUUUCCAGCCAGUCACUUGACUAAAUACUAAGCAAUUGUAUUUCUU ((((((.......((....))(((..........(((((....))))).(((((.....))))))))))))))..((((((.((....))))))))... ( -25.10) >DroSim_CAF1 3719 99 + 1 UCAAGUUUAAUCCGCCUCGGCGACACUUUUUCUUGAAGCCCGUGCUUCUUGGCUUUUCCAGCCAGUCACUUGACAAAAUACUAAGCAAUUGUAUUUCUU ((((((.......((....))(((..........(((((....))))).(((((.....))))))))))))))..((((((.((....))))))))... ( -25.20) >DroEre_CAF1 4008 99 + 1 UCAAGUUUAAUCCGCCUCGGCGACACUUUUUCUCGAAGCCCGUGCUUCUUGGCUUUUCCAGCCAGUCACUUGACAAAAUACUAAGCAAAUGUAUUUGUU ((((((.......((....))(((..........(((((....))))).(((((.....))))))))))))))..((((((.........))))))... ( -25.50) >DroYak_CAF1 4081 98 + 1 UCAAGUUUAAUCCGCCUCGGCGACACUUUUUCUUGAAGCCCGUGCUUCUUGGCUU-UCUUGCCGGUCACUUGACAAAAUACUAAGCAAAUGUAUUUGUU ((((((.......((....))(((..........(((((....))))).((((..-....)))))))))))))..((((((.........))))))... ( -24.30) >consensus UCAAGUUUAAUCCGCCUCGGCGACACUUUUUCUUGAAGCCCGUGCUUCUUGGCUUUUCCAGCCAGUCACUUGACAAAAUACUAAGCAAUUGUAUUUGUU ((((((.......((....))(((..........(((((....))))).(((((.....))))))))))))))..((((((.........))))))... (-24.34 = -24.38 + 0.04)

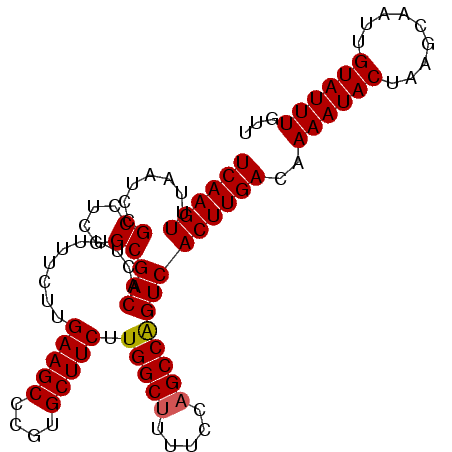

| Location | 17,486,048 – 17,486,147 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.54 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17486048 99 - 22224390 AACAAAUACAAUUGCUUAGUAUUUUGUCAAGUGACUGGCUGGAAAAGCCAAGAAGCACGGGCUUCAAGAAAAAGUGUCGCCGAGGCGGAUUAAACUUGA ...((((((.........))))))..((((((((((((((.....))))).(((((....)))))..........)))((....)).......)))))) ( -25.90) >DroSec_CAF1 3937 99 - 1 AAGAAAUACAAUUGCUUAGUAUUUAGUCAAGUGACUGGCUGGAAAAGCCAAGAAGCACGGGCUUCAAGAAAAAGUGUCGCCGAGGCGGAUUAAACUUGA ...((((((.........))))))..((((((((((((((.....))))).(((((....)))))..........)))((....)).......)))))) ( -26.60) >DroSim_CAF1 3719 99 - 1 AAGAAAUACAAUUGCUUAGUAUUUUGUCAAGUGACUGGCUGGAAAAGCCAAGAAGCACGGGCUUCAAGAAAAAGUGUCGCCGAGGCGGAUUAAACUUGA ..(((((((.........))))))).((((((((((((((.....))))).(((((....)))))..........)))((....)).......)))))) ( -27.80) >DroEre_CAF1 4008 99 - 1 AACAAAUACAUUUGCUUAGUAUUUUGUCAAGUGACUGGCUGGAAAAGCCAAGAAGCACGGGCUUCGAGAAAAAGUGUCGCCGAGGCGGAUUAAACUUGA ...((((((.........))))))..((((((((((((((.....))))).(((((....)))))..........)))((....)).......)))))) ( -25.90) >DroYak_CAF1 4081 98 - 1 AACAAAUACAUUUGCUUAGUAUUUUGUCAAGUGACCGGCAAGA-AAGCCAAGAAGCACGGGCUUCAAGAAAAAGUGUCGCCGAGGCGGAUUAAACUUGA ...((((((.........))))))..(((((((((((((....-..)))..(((((....)))))........).)))((....)).......)))))) ( -23.80) >consensus AACAAAUACAAUUGCUUAGUAUUUUGUCAAGUGACUGGCUGGAAAAGCCAAGAAGCACGGGCUUCAAGAAAAAGUGUCGCCGAGGCGGAUUAAACUUGA ...((((((.........))))))..(((((((((.((((.....))))..(((((....)))))..........)))((....)).......)))))) (-24.34 = -24.54 + 0.20)


| Location | 17,486,074 – 17,486,179 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 95.43 |
| Mean single sequence MFE | -20.06 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.50 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.999066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17486074 105 + 22224390 UUUUUCUUGAAGCCCGUGCUUCUUGGCUUUUCCAGCCAGUCACUUGACAAAAUACUAAGCAAUUGUAUUUGUUGUAUUUGCUUUAAUAAUUAUUUUCCCACCAUG ......(((((((..((((....(((((.....)))))(((....))).((((((.((....))))))))...))))..)))))))................... ( -20.50) >DroSec_CAF1 3963 105 + 1 UUUUUCUUGAAGCCCGUGCUUCUUGGCUUUUCCAGCCAGUCACUUGACUAAAUACUAAGCAAUUGUAUUUCUUGUAUUUGCUUUAAUAAUUAUUUUCCCACCACG ......(((((((..((((.....((((.....))))((((....))))((((((.((....))))))))...))))..)))))))................... ( -19.60) >DroSim_CAF1 3745 105 + 1 UUUUUCUUGAAGCCCGUGCUUCUUGGCUUUUCCAGCCAGUCACUUGACAAAAUACUAAGCAAUUGUAUUUCUUGUAUUUGCUUUAAUAAUUAUUUUCCCACCACG ......(((((((..((((....(((((.....)))))(((....))).((((((.((....))))))))...))))..)))))))................... ( -19.90) >DroEre_CAF1 4034 105 + 1 UUUUUCUCGAAGCCCGUGCUUCUUGGCUUUUCCAGCCAGUCACUUGACAAAAUACUAAGCAAAUGUAUUUGUUGUAUUUGCUUUAAUAAUUAUUUUCCCACCACG ........(((((....))))).(((((.....)))))(((....)))........(((((((((((.....)))))))))))...................... ( -21.50) >DroYak_CAF1 4107 104 + 1 UUUUUCUUGAAGCCCGUGCUUCUUGGCUU-UCUUGCCGGUCACUUGACAAAAUACUAAGCAAAUGUAUUUGUUGUAUUUGUUUCAAUAAUUAUUUUCCCACCACG ......(((((((..((((....((((..-....))))(((....))).((((((.........))))))...))))..)))))))................... ( -18.80) >consensus UUUUUCUUGAAGCCCGUGCUUCUUGGCUUUUCCAGCCAGUCACUUGACAAAAUACUAAGCAAUUGUAUUUGUUGUAUUUGCUUUAAUAAUUAUUUUCCCACCACG ......(((((((..((((....(((((.....)))))(((....))).((((((.........))))))...))))..)))))))................... (-18.58 = -18.50 + -0.08)



| Location | 17,486,074 – 17,486,179 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 95.43 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -17.24 |
| Energy contribution | -17.84 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17486074 105 - 22224390 CAUGGUGGGAAAAUAAUUAUUAAAGCAAAUACAACAAAUACAAUUGCUUAGUAUUUUGUCAAGUGACUGGCUGGAAAAGCCAAGAAGCACGGGCUUCAAGAAAAA (((..(((.((((((.......((((((...............))))))..)))))).))).)))..(((((.....))))).(((((....)))))........ ( -22.96) >DroSec_CAF1 3963 105 - 1 CGUGGUGGGAAAAUAAUUAUUAAAGCAAAUACAAGAAAUACAAUUGCUUAGUAUUUAGUCAAGUGACUGGCUGGAAAAGCCAAGAAGCACGGGCUUCAAGAAAAA .((..(..((...(((.((((((.((((...............)))))))))).))).))..)..))(((((.....))))).(((((....)))))........ ( -21.66) >DroSim_CAF1 3745 105 - 1 CGUGGUGGGAAAAUAAUUAUUAAAGCAAAUACAAGAAAUACAAUUGCUUAGUAUUUUGUCAAGUGACUGGCUGGAAAAGCCAAGAAGCACGGGCUUCAAGAAAAA .....(((.((((((.......((((((...............))))))..)))))).)))......(((((.....))))).(((((....)))))........ ( -20.76) >DroEre_CAF1 4034 105 - 1 CGUGGUGGGAAAAUAAUUAUUAAAGCAAAUACAACAAAUACAUUUGCUUAGUAUUUUGUCAAGUGACUGGCUGGAAAAGCCAAGAAGCACGGGCUUCGAGAAAAA .................((((((.((((((...........))))))))))))((((.((.......(((((.....))))).(((((....))))))).)))). ( -23.30) >DroYak_CAF1 4107 104 - 1 CGUGGUGGGAAAAUAAUUAUUGAAACAAAUACAACAAAUACAUUUGCUUAGUAUUUUGUCAAGUGACCGGCAAGA-AAGCCAAGAAGCACGGGCUUCAAGAAAAA ...(((.(...........((((...((((((((((((....)))).)).))))))..)))).).)))(((....-..)))..(((((....)))))........ ( -18.30) >consensus CGUGGUGGGAAAAUAAUUAUUAAAGCAAAUACAACAAAUACAAUUGCUUAGUAUUUUGUCAAGUGACUGGCUGGAAAAGCCAAGAAGCACGGGCUUCAAGAAAAA .....(((.((((((.......((((((...............))))))..)))))).))).......((((.....))))..(((((....)))))........ (-17.24 = -17.84 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:25 2006