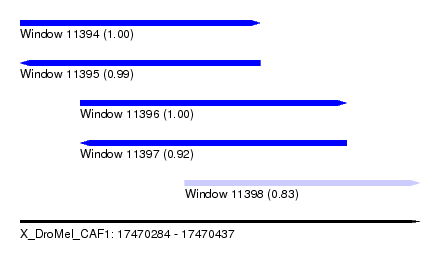
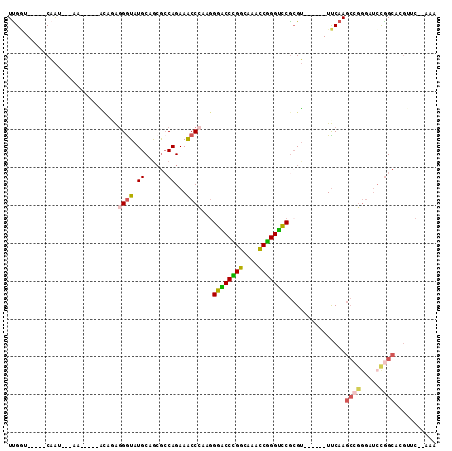
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,470,284 – 17,470,437 |
| Length | 153 |
| Max. P | 0.999519 |

| Location | 17,470,284 – 17,470,376 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.20 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -22.35 |
| Energy contribution | -22.18 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17470284 92 + 22224390 UUGGU-----CAAU---AA-----ACAGAGGGUAUGCAGCGCCAGAAACCCAAGGGACCCGGCAAACCGGGUUCGCGU------UUCAAGCCGGGAUCCGGCACGUUC--AAA .....-----....---..-----...(((.((.((.(((((..(.....)...((((((((....))))))))))))------).)).(((((...))))))).)))--... ( -31.90) >DroSec_CAF1 428 93 + 1 UUGGU-----CAAAA--AA-----ACAGAGGGUAUGCAGCGCCAGAAACCCAAGGGACCCGGCAAACCGGGUCCGCGU------UUCAAGCCGGGUUCCGGCACGUUC--AAA .....-----.....--..-----...(((.((.((.(((((..(.....)...((((((((....))))))))))))------).)).(((((...))))))).)))--... ( -34.80) >DroSim_CAF1 53 95 + 1 UUGGU-----CAAAAAAAA-----ACAGAGGGUAUGCAGCGCCAGAAACCCAAGGGACCCGGCAAACCGGGUCCGCGU------UUCAAGCCGGGUUCCGGCACGUUC--AAA .....-----.........-----...(((.((.((.(((((..(.....)...((((((((....))))))))))))------).)).(((((...))))))).)))--... ( -34.80) >DroEre_CAF1 27 92 + 1 UCGAC-----CAAU---AA-----ACAAAGGGUAUGCAGCGCCAGAAACCCAAGGGACCCGGCAAAUCGGGUCCGCGU------UUCAAGCCGGGAUCUGGCAAGAGC--AAA ...((-----(...---..-----......))).(((...((((((..(((...((((((((....))))))))((..------.....)).))).))))))....))--).. ( -33.80) >DroMoj_CAF1 2085 106 + 1 UUGCUUUUAACAAUUACAAAUAAAGUACGCGGUAUGCAAAAACAGAAGCCUCGAGGUCCUGGAAAACCAGGAUCGCGUGCGCCAUCCAAGCCGGGCGGCAGCC---AA--A-- ..(((...................(((((((((.((......))...)))....((((((((....))))))))))))))(((..((.....))..)))))).---..--.-- ( -36.50) >DroAna_CAF1 678 88 + 1 UUGAA-----CCGC---AU-----UAACAGGAUAUGCAGCGCCAGAAACCAAGGGGGCCGGGCAAACCCGGUCCAAGG------UUCAAA------UCCGGCAAGGGCUUCAA .((((-----((((---((-----.........))))...(((.((((((....((((((((....))))))))..))------))....------)).)))...)).)))). ( -33.40) >consensus UUGGU_____CAAU___AA_____ACAGAGGGUAUGCAGCGCCAGAAACCCAAGGGACCCGGCAAACCGGGUCCGCGU______UUCAAGCCGGGAUCCGGCACGUUC__AAA .............................((((.((......))...))))...((((((((....))))))))...............((((.....))))........... (-22.35 = -22.18 + -0.16)


| Location | 17,470,284 – 17,470,376 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 69.20 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -21.83 |
| Energy contribution | -22.25 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17470284 92 - 22224390 UUU--GAACGUGCCGGAUCCCGGCUUGAA------ACGCGAACCCGGUUUGCCGGGUCCCUUGGGUUUCUGGCGCUGCAUACCCUCUGU-----UU---AUUG-----ACCAA ...--....((((((((.((((((.....------..))(.((((((....)))))).)..))))..))))))))..............-----..---....-----..... ( -31.60) >DroSec_CAF1 428 93 - 1 UUU--GAACGUGCCGGAACCCGGCUUGAA------ACGCGGACCCGGUUUGCCGGGUCCCUUGGGUUUCUGGCGCUGCAUACCCUCUGU-----UU--UUUUG-----ACCAA ...--....(((((((((((((((.....------..))((((((((....))))))))..)))).)))))))))..............-----..--.....-----..... ( -39.70) >DroSim_CAF1 53 95 - 1 UUU--GAACGUGCCGGAACCCGGCUUGAA------ACGCGGACCCGGUUUGCCGGGUCCCUUGGGUUUCUGGCGCUGCAUACCCUCUGU-----UUUUUUUUG-----ACCAA ...--....(((((((((((((((.....------..))((((((((....))))))))..)))).)))))))))..............-----.........-----..... ( -39.70) >DroEre_CAF1 27 92 - 1 UUU--GCUCUUGCCAGAUCCCGGCUUGAA------ACGCGGACCCGAUUUGCCGGGUCCCUUGGGUUUCUGGCGCUGCAUACCCUUUGU-----UU---AUUG-----GUCGA ..(--((...(((((((.((((((.....------..))(((((((......)))))))..))))..)))))))..)))..........-----..---....-----..... ( -34.80) >DroMoj_CAF1 2085 106 - 1 --U--UU---GGCUGCCGCCCGGCUUGGAUGGCGCACGCGAUCCUGGUUUUCCAGGACCUCGAGGCUUCUGUUUUUGCAUACCGCGUACUUUAUUUGUAAUUGUUAAAAGCAA --.--..---.(((((((.((.....)).))))..(((((.((((((....))))))....(((((....))))).......))))).....................))).. ( -28.90) >DroAna_CAF1 678 88 - 1 UUGAAGCCCUUGCCGGA------UUUGAA------CCUUGGACCGGGUUUGCCCGGCCCCCUUGGUUUCUGGCGCUGCAUAUCCUGUUA-----AU---GCGG-----UUCAA .(((.......((((((------....((------((..((.(((((....)))))...))..))))))))))(((((((.........-----))---))))-----)))). ( -33.50) >consensus UUU__GAACGUGCCGGAACCCGGCUUGAA______ACGCGGACCCGGUUUGCCGGGUCCCUUGGGUUUCUGGCGCUGCAUACCCUCUGU_____UU___AUUG_____ACCAA ..........(((((((..((.((.............))((((((((....))))))))....))..)))))))....................................... (-21.83 = -22.25 + 0.42)
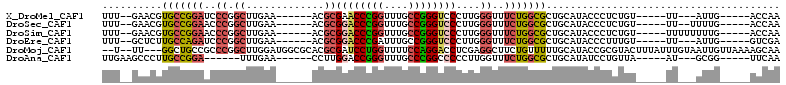
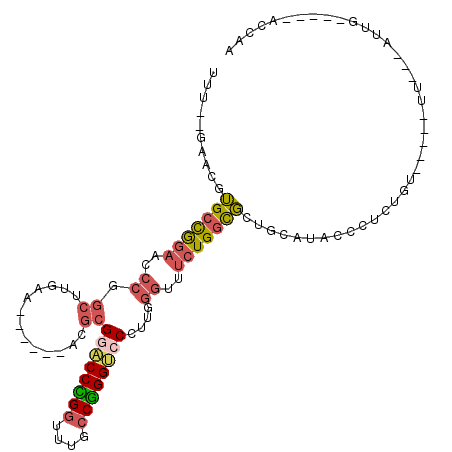
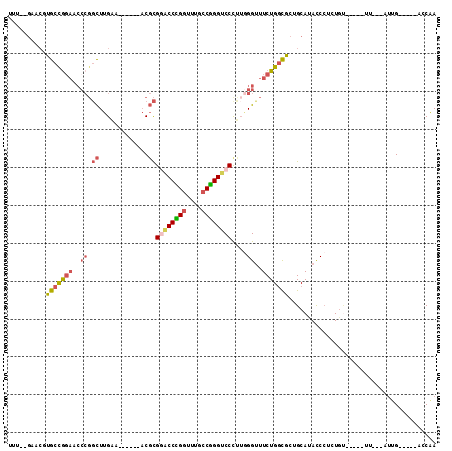
| Location | 17,470,307 – 17,470,409 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -44.88 |
| Consensus MFE | -30.08 |
| Energy contribution | -32.32 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.67 |
| SVM decision value | 3.68 |
| SVM RNA-class probability | 0.999519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17470307 102 + 22224390 CAGCGCCAGAAACCCAAGGGACCCGGCAAACCGGGUUCGCGUUUCA---AGCCGGGAUCCGGCACGUUCAAAGGAGCUGGUGGUGGCCGGAAAAGCGGCGACAAC ((((.((.(((((.(...((((((((....))))))))).))))).---.(((((...))))).........)).))))...((.((((......)))).))... ( -45.60) >DroSec_CAF1 452 102 + 1 CAGCGCCAGAAACCCAAGGGACCCGGCAAACCGGGUCCGCGUUUCA---AGCCGGGUUCCGGCACGUUCAAAGGAGCUGCUGGUGGGCGGAAAAGCGGCGACAAC ..(((((...........((((((((....))))))))((.((((.---.(((.(...((((((.((((....))))))))))).))).)))).)))))).)... ( -48.10) >DroSim_CAF1 79 102 + 1 CAGCGCCAGAAACCCAAGGGACCCGGCAAACCGGGUCCGCGUUUCA---AGCCGGGUUCCGGCACGUUCAAAGGAGCUGCUGGUGGCCGGAAAAGCGGCGACAAC (((((((.(((.(((...((((((((....))))))))((......---.)).)))))).)))..((((....)))).))))((.((((......)))).))... ( -51.00) >DroEre_CAF1 50 102 + 1 CAGCGCCAGAAACCCAAGGGACCCGGCAAAUCGGGUCCGCGUUUCA---AGCCGGGAUCUGGCAAGAGCAAAGGAGCUCCUGGUGGCCGGAAGAGUGGCGACCAC ....((((((..(((...((((((((....))))))))((......---.)).))).))))))..((((......)))).((((.((((......)))).)))). ( -48.20) >DroWil_CAF1 655 93 + 1 CAGAGGCAUAAACCAAAGCGUCCUGGU---CCUGGUGCUGGUCUUAGAAAGCCAAA---CGGCG---UAAAGGGACCGGCUGGC---AAGGACAAAUUCAACAAA (((.(((............))))))((---(((.(((((((((((.....(((...---.))).---....)))))))))..))---.)))))............ ( -31.50) >consensus CAGCGCCAGAAACCCAAGGGACCCGGCAAACCGGGUCCGCGUUUCA___AGCCGGGAUCCGGCACGUUCAAAGGAGCUGCUGGUGGCCGGAAAAGCGGCGACAAC ((((.((.(((((.(...((((((((....))))))))).))))).....(((((...))))).........)).))))...((.((((......)))).))... (-30.08 = -32.32 + 2.24)

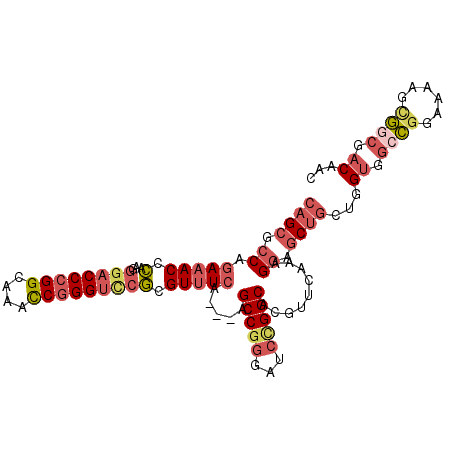
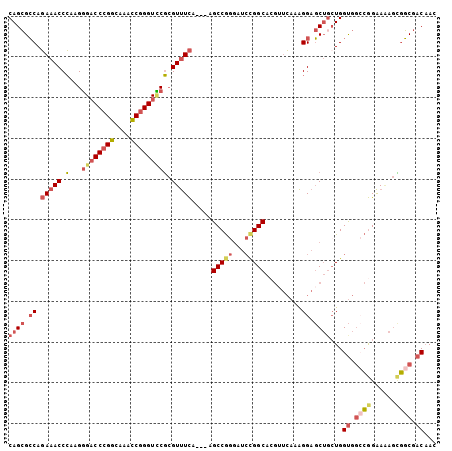
| Location | 17,470,307 – 17,470,409 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -41.13 |
| Consensus MFE | -27.20 |
| Energy contribution | -28.60 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17470307 102 - 22224390 GUUGUCGCCGCUUUUCCGGCCACCACCAGCUCCUUUGAACGUGCCGGAUCCCGGCU---UGAAACGCGAACCCGGUUUGCCGGGUCCCUUGGGUUUCUGGCGCUG ((.((.((((......)))).)).))((((.((.........(((((...))))).---.(((((.(((((((((....))))))...))).))))).)).)))) ( -42.00) >DroSec_CAF1 452 102 - 1 GUUGUCGCCGCUUUUCCGCCCACCAGCAGCUCCUUUGAACGUGCCGGAACCCGGCU---UGAAACGCGGACCCGGUUUGCCGGGUCCCUUGGGUUUCUGGCGCUG (((((.(..((......))....).)))))..........(((((((((((((((.---......))((((((((....))))))))..)))).))))))))).. ( -43.90) >DroSim_CAF1 79 102 - 1 GUUGUCGCCGCUUUUCCGGCCACCAGCAGCUCCUUUGAACGUGCCGGAACCCGGCU---UGAAACGCGGACCCGGUUUGCCGGGUCCCUUGGGUUUCUGGCGCUG ((((..((((......))))...)))).............(((((((((((((((.---......))((((((((....))))))))..)))).))))))))).. ( -48.70) >DroEre_CAF1 50 102 - 1 GUGGUCGCCACUCUUCCGGCCACCAGGAGCUCCUUUGCUCUUGCCAGAUCCCGGCU---UGAAACGCGGACCCGAUUUGCCGGGUCCCUUGGGUUUCUGGCGCUG (((((((.........))))))).((((((......))))))((((((.((((((.---......))(((((((......)))))))..))))..)))))).... ( -48.40) >DroWil_CAF1 655 93 - 1 UUUGUUGAAUUUGUCCUU---GCCAGCCGGUCCCUUUA---CGCCG---UUUGGCUUUCUAAGACCAGCACCAGG---ACCAGGACGCUUUGGUUUAUGCCUCUG ............(((((.---(((.((.((((......---.(((.---...))).......)))).))....))---.).))))).....(((....))).... ( -22.64) >consensus GUUGUCGCCGCUUUUCCGGCCACCAGCAGCUCCUUUGAACGUGCCGGAACCCGGCU___UGAAACGCGGACCCGGUUUGCCGGGUCCCUUGGGUUUCUGGCGCUG ...((.(((........))).))...((((.((.........((((.....)))).....(((((.(((((((((....))))))))...).))))).)).)))) (-27.20 = -28.60 + 1.40)


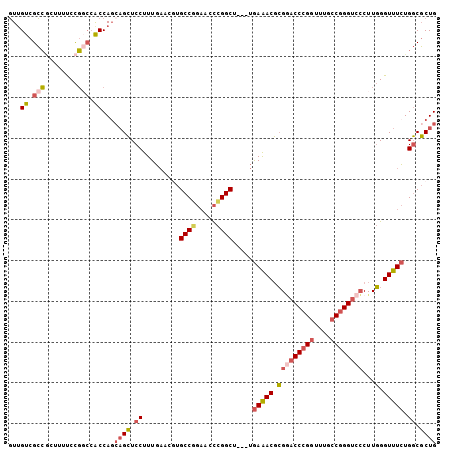
| Location | 17,470,347 – 17,470,437 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.74 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17470347 90 + 22224390 GUUUCAAGCCGGGAUCCGGCACGUUCAAAGGAGCUGGUGGUGGCCGGAAAAGCGGCGACAACAGCCAGAAGCGUCCGCGCCAGGAGAUCA (((((..(((((...)))))..((((....))))((((.((.((((......)))).))....))))))))).(((......)))..... ( -34.20) >DroSec_CAF1 492 90 + 1 GUUUCAAGCCGGGUUCCGGCACGUUCAAAGGAGCUGCUGGUGGGCGGAAAAGCGGCGACAACAGCCAAAAGCGUCCGCGCCAGGAGAUCA (((((..(((((...)))))..((((....))))..(((((..(((((...(((((.......)))....)).))))))))))))))).. ( -34.70) >DroSim_CAF1 119 90 + 1 GUUUCAAGCCGGGUUCCGGCACGUUCAAAGGAGCUGCUGGUGGCCGGAAAAGCGGCGACAACAGCCAAAAGCGUCCGCGCCAGGAGAUCA (((((...((((.(.((((((.((((....)))))))))).).))))......((((...((.((.....))))...))))..))))).. ( -33.60) >DroEre_CAF1 90 90 + 1 GUUUCAAGCCGGGAUCUGGCAAGAGCAAAGGAGCUCCUGGUGGCCGGAAGAGUGGCGACCACAACCAAAAGCGUCCGCGCCAGGAGCUUA ((((...(((((...)))))..))))....((((((((((((..((((.(..(((.........)))....).)))))))))))))))). ( -37.00) >DroYak_CAF1 116 90 + 1 GUUUCAAGCCGGGCUCCGGCAAGAACAAAGGAGCUGCUGGUGGCCGAAAAAGUGGCGACAACAACCAGAAACGUCCGCGCCAGGAGCUCA ((((...(((((...)))))..))))....(((((.((((((((((......))))(((.............)))..)))))).))))). ( -35.92) >consensus GUUUCAAGCCGGGAUCCGGCACGUUCAAAGGAGCUGCUGGUGGCCGGAAAAGCGGCGACAACAGCCAAAAGCGUCCGCGCCAGGAGAUCA .......(((((...)))))..........((.((.((((((..((((...(((((.......)))....)).)))))))))).)).)). (-26.66 = -26.74 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:11 2006