
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,464,396 – 17,464,538 |
| Length | 142 |
| Max. P | 0.996774 |

| Location | 17,464,396 – 17,464,510 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -28.44 |
| Consensus MFE | -14.60 |
| Energy contribution | -18.72 |
| Covariance contribution | 4.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17464396 114 + 22224390 CAUCAGUCUUCGGGUUUUCGCCGAUUUUUACUUGUUUGGCCGCAUCAG-CCGGC-----UCCUCCUCUCCUGAAGCCUCCCCUCCGGAUUCCUUGUCAUCCGAUACGGAUUCCUCGACUU ....((((...(((.....(((((...........))))).((.((((-..((.-----....))....)))).))...)))...(((.(((.((((....)))).))).)))..)))). ( -29.40) >DroSec_CAF1 6606 114 + 1 CAUCAGGUUGCGGUUUUUCGCCGAAUUUCACUUGUUUGGCCGCAUCAG-CCGGC-----UCUUCCUCUCCUGAUGCCUCGCCUCCUGAUUCCUUGUCAUCCGAUACGGAUUCCUCCACUU .((((((..((((......(((((((.......))))))).(((((((-..((.-----....))....))))))).))))..))))))(((.((((....)))).)))........... ( -36.40) >DroSim_CAF1 6715 114 + 1 CAUCAGGCUGCGGUUUUUCGCCGAAUUUCACUUGUUUGGCCGCAUCAG-CCGGC-----UCUUCCUCUCCUGAUGCCUCGCCUCCUGAUUCCUUGUCAUCCGAUACGGAUUCCUCCACUU .((((((..((((......(((((((.......))))))).(((((((-..((.-----....))....))))))).))))..))))))(((.((((....)))).)))........... ( -35.40) >DroAna_CAF1 8294 111 + 1 CAUCAGACUUC-----UUCUCC----UCAACCUCUUUGGAACUAUCGGGCUGGCACACAUCUUCCUCUCCUGCCUUAUCCUUCUCAGAUUUCUUGGCAACUACAAUCGAAUCAUCUUCAU .....((.(((-----......----..........(((((..((.((((.((...............)).)))).))..))).))((((....(....)...)))))))))........ ( -12.56) >consensus CAUCAGGCUGCGGUUUUUCGCCGAAUUUCACUUGUUUGGCCGCAUCAG_CCGGC_____UCUUCCUCUCCUGAUGCCUCCCCUCCUGAUUCCUUGUCAUCCGAUACGGAUUCCUCCACUU .((((((..((((......(((((((.......))))))).(((((((...((..........))....))))))).))))..))))))(((.((((....)))).)))........... (-14.60 = -18.72 + 4.12)

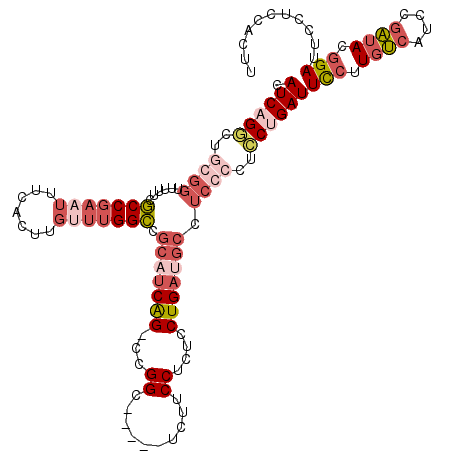
| Location | 17,464,436 – 17,464,538 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.43 |
| Mean single sequence MFE | -19.21 |
| Consensus MFE | -6.92 |
| Energy contribution | -8.70 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.36 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17464436 102 + 22224390 CGCAUCAG-CCGGC-----UCCUCCUCUCCUGAAGCCUCCCCUCCGGAUUCCUUGUCAUCCGAUACGGAUUCCUCGACUUCUGAGCCA------------GAUUUCUCAUCAUAUCUAUC .((.((((-..(((-----(.(.........).))))........(((.(((.((((....)))).))).))).......)))))).(------------(((..........))))... ( -21.80) >DroSec_CAF1 6646 102 + 1 CGCAUCAG-CCGGC-----UCUUCCUCUCCUGAUGCCUCGCCUCCUGAUUCCUUGUCAUCCGAUACGGAUUCCUCCACUUCUGAGCCA------------GAUUUAUCAUCAUAUCUGUC .(((((((-..((.-----....))....)))))))((((......((.(((.((((....)))).))).)).........)))).((------------(((..........))))).. ( -20.36) >DroSim_CAF1 6755 102 + 1 CGCAUCAG-CCGGC-----UCUUCCUCUCCUGAUGCCUCGCCUCCUGAUUCCUUGUCAUCCGAUACGGAUUCCUCCACUUCUGAGCCA------------GAUUUAUCAUCAUAUCUGUC .(((((((-..((.-----....))....)))))))((((......((.(((.((((....)))).))).)).........)))).((------------(((..........))))).. ( -20.36) >DroEre_CAF1 14687 102 + 1 CACAUCAU-CCGGC-----UCCUCCUCUCCUGAUGCCUCUCCUCCAGAUUCCUUGUAGUCCGAUACGGAUGCCUCCAUUUCUGAUCCA------------GAUUUCUCAUCAUAUCCGUC ......((-(.(((-----(.(.......(((..(......)..))).......).)))).)))(((((((........((((...))------------))..........))))))). ( -13.91) >DroYak_CAF1 6759 102 + 1 CGCAUCAU-CCGAC-----UCCUCCUCUCCUGAUGCUUCCUCUCCUGAUUCCUUGUCGUCCGAUAGGGAUUCCUCCAUUUCUGAUCCA------------GAUUUCUCAUCAUAUUCGCC .((((((.-..((.-----......))...))))))..........((.((((((((....)))))))).)).......((((...))------------)).................. ( -19.40) >DroAna_CAF1 8325 120 + 1 ACUAUCGGGCUGGCACACAUCUUCCUCUCCUGCCUUAUCCUUCUCAGAUUUCUUGGCAACUACAAUCGAAUCAUCUUCAUCUGAAUCAUCUGACUCUGAAGAUUCCUCCUCCGAAUUGUC ....(((((..((((...............)))).....((((((((((.....(....).......((.(((........))).))))))))....))))........)))))...... ( -19.46) >consensus CGCAUCAG_CCGGC_____UCCUCCUCUCCUGAUGCCUCCCCUCCUGAUUCCUUGUCAUCCGAUACGGAUUCCUCCACUUCUGAGCCA____________GAUUUCUCAUCAUAUCUGUC .(((((((...((..........))....)))))))..........((.(((.((((....)))).))).))................................................ ( -6.92 = -8.70 + 1.78)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:05 2006