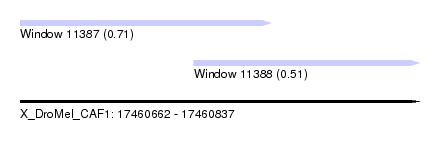
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,460,662 – 17,460,837 |
| Length | 175 |
| Max. P | 0.709979 |

| Location | 17,460,662 – 17,460,772 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.96 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -19.50 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17460662 110 + 22224390 AUUAGGUAAAUGACUACCUAGCAUAUCUAAUCAUAGUUGGAUGUAGCCUAGCUAUUUUAAGCGGAUCAUUGGUCGACAUCUCUGCUCGCAAAUCAAGCGAGACAAGCUAA---- ..((((((......))))))(((((((((((....))))))))).)).(((((.......(((((....((.....))..)))))((((.......))))....))))).---- ( -29.70) >DroSec_CAF1 2942 110 + 1 AUUAGAUAAAUGACUACCUGGCAUAUCUAAUCAUAGUUGGAUAUGGACUAGCUAUUUUAAGCGGAUCAUUGGUCGACAUCUCUGCUCGCAAAUCAAGCGAGACAAGCAAG---- ....(((.(((((...((...((((((((((....)))))))))).....(((......))))).))))).)))........(((((((.......))))).))......---- ( -27.30) >DroSim_CAF1 3044 88 + 1 ----------------------AUAUCUAAUCAUAGUUGGAUAUGGUCUAGCUAUUUUAAGCGGAUCAUUGGUCGACAUCUCUGCUCGCAAAUCAAGCGAGACAAGCAAG---- ----------------------.............(((((..(((((((.(((......))))))))))...))))).....(((((((.......))))).))......---- ( -23.20) >DroEre_CAF1 10958 111 + 1 -CUAGAUAAAUGACUGCCUGGCAUAUCUAAUCAUAGUUGGAUGUGGUCUAGCUAUUUUAAGCGGAUCUUUGGCCGACAUCUCUGCUCGCAAAUCAUGCGAGACAAACUGGCA-- -..............((((((((((((((((....)))))))))(((((.(((......))))))))....)))).......((((((((.....)))))).))....))).-- ( -33.00) >DroYak_CAF1 3021 114 + 1 ACUAGAUAAAUAACUACCUGGCAUAUCUAAUCAUAGUUGGAUGUGCUCUAGCUAUUUUAAGCGGAUCUUUGGCCGACAUCUCUGCUCGCAAAUCAAGCGAGGCAAACUGACAAG ...(((((((((.(((...((((((((((((....)))))))))))).))).)))))...((((........))).))))).(((((((.......)))).))).......... ( -27.90) >consensus ACUAGAUAAAUGACUACCUGGCAUAUCUAAUCAUAGUUGGAUGUGGUCUAGCUAUUUUAAGCGGAUCAUUGGUCGACAUCUCUGCUCGCAAAUCAAGCGAGACAAGCUAG____ ...................................(((((..(((((((.(((......))))))))))...))))).....(((((((.......))))).)).......... (-19.50 = -20.22 + 0.72)



| Location | 17,460,738 – 17,460,837 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 86.77 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -17.50 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17460738 99 + 22224390 CAUCUCUGCUCGCAAAUCAAGCGAGACAAGCUAA-----------GCAUACACACACACACACAUGGAGGCCACUUAACACCUUACAGGUGCACGAAUGCGCAGGGGACA ..(((((((((((.......))))..........-----------((((.(...............(((....)))..((((.....))))...).)))))))))))... ( -24.60) >DroSec_CAF1 3018 97 + 1 CAUCUCUGCUCGCAAAUCAAGCGAGACAAGCAAG-----------GCAUACACAC--ACACACAUGGAGGCCACUUAACACCUUACAGGUGCACGAAUGCGCAGGGGACA ..(((((((((((.......)))).....(((.(-----------((...((...--.......))...)))......((((.....))))......))))))))))... ( -26.90) >DroSim_CAF1 3098 97 + 1 CAUCUCUGCUCGCAAAUCAAGCGAGACAAGCAAG-----------GCAUACACAC--ACACACAUGGAGGCCACUUAACACCUUACAGGUGCACGAAUGCGCAGGGGACA ..(((((((((((.......)))).....(((.(-----------((...((...--.......))...)))......((((.....))))......))))))))))... ( -26.90) >DroEre_CAF1 11033 94 + 1 CAUCUCUGCUCGCAAAUCAUGCGAGACAAACUGGCA--------------CACAC--ACACACAUAGAUGCCACUUAACACCUCACAGGUGCACGAAUGCGCUGGGGACA ......((((((((.....)))))).))...(((((--------------(....--.........).))))).......(((((...(((((....))))))))))... ( -24.12) >DroYak_CAF1 3097 108 + 1 CAUCUCUGCUCGCAAAUCAAGCGAGGCAAACUGACAAGCAGACACACAUGCACAC--ACACACAUGGAGGCCACUUAACACCUUACAGGUGCACGAAUGCGAUGGGGACA ..((((((.(((((..........(((...(((.....))).....((((.....--.....))))...)))......((((.....))))......))))))))))).. ( -25.50) >consensus CAUCUCUGCUCGCAAAUCAAGCGAGACAAGCUAG___________GCAUACACAC__ACACACAUGGAGGCCACUUAACACCUUACAGGUGCACGAAUGCGCAGGGGACA ..(((((.(((((.......)))))............................................((.......((((.....)))).......))...))))).. (-17.50 = -17.50 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:27:02 2006