
| Sequence ID | X_DroMel_CAF1 |
|---|---|
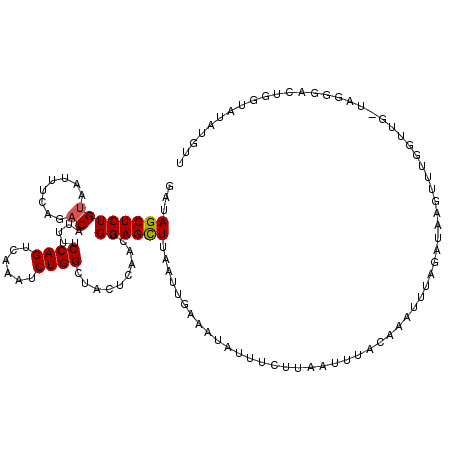
| Location | 17,459,737 – 17,459,857 |
| Length | 120 |
| Max. P | 0.945297 |

| Location | 17,459,737 – 17,459,857 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.76 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -9.78 |
| Energy contribution | -9.82 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17459737 120 + 22224390 GAUAGCUCUGUAAUUUCAGCCAUUUUCCAGUCAAAUCUGGCUACUCAACUGGAGCUUAAUUGAAAUAUUUCUUAAUUUACAAAUUUAGAUAAGUUUGGUUGCUAGGGACUGGUAUAUAAU .......(((......))).......((((((...((((((.(((.((((....((.((((.((((........))))...)))).))...)))).))).))))))))))))........ ( -24.60) >DroSec_CAF1 2037 120 + 1 GAUAGCUCUGUAAUUUCAGACAUUUUCCAGUCAAAACUGGCUACUCAACUGGAGCUUAAUUGAAAUAUUUCUUAAUUUACAAAUUUAGAUAAGUUUGGUUGAUAGGGACUGGUAUAUGUU ......((((......))))(((...((((((....(((.....(((((..((.((((.(((((...................))))).))))))..)))))))).))))))...))).. ( -25.21) >DroSim_CAF1 2155 120 + 1 GAUAGCUCUGUAAUUUCAGACAUUUUCCAGUCAAAACUGGCUACUCAACUGGAGCUUAAUUGAAAUAUUUCUUAAUAUACAAAUUUAGAUAAGUUUGGUUGAUAGGGACUAUUAUAUGUU (((((((((((...............(((((....))))).....((((..((.((((.((((((((((....))))).....))))).))))))..)))))))))).)))))....... ( -25.10) >DroEre_CAF1 10065 94 + 1 GAUAGCUCUGUAAUUUCAGACAUUUUCCAGUCAAAUCUGGCUAUUCAACUGGAGCUUGGCUUAAGCAUUUCAAAACUGGA--------------------------AAAUGGUAUAGUUU ......((((......))))(((((((((((....((..(........)..))(((((...)))))........))))))--------------------------)))))......... ( -23.70) >DroYak_CAF1 2208 92 + 1 GAUAGCUCUGUAACUUCAGACAUUUUCCAGUCAAAUCUGGCUAUUUAACUGGAGUUUAAUGCAAAUGUUUCUAAAUUUGC----------------------------AUGGGCUAGUUA ..((((((...((((((((.......((((......))))........))))))))..((((((((........))))))----------------------------)))))))).... ( -27.66) >consensus GAUAGCUCUGUAAUUUCAGACAUUUUCCAGUCAAAUCUGGCUACUCAACUGGAGCUUAAUUGAAAUAUUUCUUAAUUUACAAAUUUAGAUAAGUUUGGUUG_UAGGGACUGGUAUAUGUU ...((((((((........)).....((((......))))..........))))))................................................................ ( -9.78 = -9.82 + 0.04)


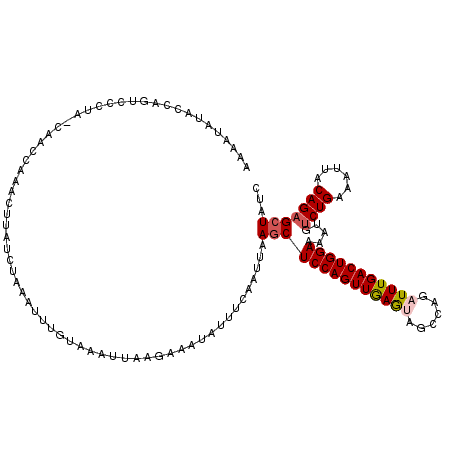
| Location | 17,459,737 – 17,459,857 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.76 |
| Mean single sequence MFE | -20.76 |
| Consensus MFE | -14.80 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17459737 120 - 22224390 AUUAUAUACCAGUCCCUAGCAACCAAACUUAUCUAAAUUUGUAAAUUAAGAAAUAUUUCAAUUAAGCUCCAGUUGAGUAGCCAGAUUUGACUGGAAAAUGGCUGAAAUUACAGAGCUAUC ................((((..(((......(((.((((....)))).)))................((((((..(((......)))..))))))...)))(((......))).)))).. ( -21.40) >DroSec_CAF1 2037 120 - 1 AACAUAUACCAGUCCCUAUCAACCAAACUUAUCUAAAUUUGUAAAUUAAGAAAUAUUUCAAUUAAGCUCCAGUUGAGUAGCCAGUUUUGACUGGAAAAUGUCUGAAAUUACAGAGCUAUC ...............................(((.((((....)))).))).............(((((((((..((........))..)))))).....((((......)))))))... ( -18.10) >DroSim_CAF1 2155 120 - 1 AACAUAUAAUAGUCCCUAUCAACCAAACUUAUCUAAAUUUGUAUAUUAAGAAAUAUUUCAAUUAAGCUCCAGUUGAGUAGCCAGUUUUGACUGGAAAAUGUCUGAAAUUACAGAGCUAUC ........((((..(................(((.(((......))).)))...((((((..((...((((((..((........))..))))))...))..))))))....)..)))). ( -18.00) >DroEre_CAF1 10065 94 - 1 AAACUAUACCAUUU--------------------------UCCAGUUUUGAAAUGCUUAAGCCAAGCUCCAGUUGAAUAGCCAGAUUUGACUGGAAAAUGUCUGAAAUUACAGAGCUAUC ..............--------------------------...(((((((....((((.....))))((((((..(((......)))..))))))...............)))))))... ( -21.60) >DroYak_CAF1 2208 92 - 1 UAACUAGCCCAU----------------------------GCAAAUUUAGAAACAUUUGCAUUAAACUCCAGUUAAAUAGCCAGAUUUGACUGGAAAAUGUCUGAAGUUACAGAGCUAUC ....((((..((----------------------------((((((........)))))))).....(((((((((((......))))))))))).....((((......)))))))).. ( -24.70) >consensus AAAAUAUACCAGUCCCUA_CAACCAAACUUAUCUAAAUUUGUAAAUUAAGAAAUAUUUCAAUUAAGCUCCAGUUGAGUAGCCAGAUUUGACUGGAAAAUGUCUGAAAUUACAGAGCUAUC ................................................................((((((((((((((......))))))))))).....((((......)))))))... (-14.80 = -15.20 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:59 2006