
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,442,987 – 17,443,132 |
| Length | 145 |
| Max. P | 0.996591 |

| Location | 17,442,987 – 17,443,093 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -21.11 |
| Energy contribution | -22.87 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17442987 106 + 22224390 ----AGCACAC---AGCAACUGCAAAGUUAAUCAAAAUGCAAUGUGGUCAAAAGGGCGUGAACA-GGCCAGGUGCACGUCAUAUUCGCACCUUUUGUAAACGACGCCCUUCC------CU ----..((((.---.((((((....))).........)))..)))).....(((((((((.(((-((..((((((...........)))))))))))...).))))))))..------.. ( -27.90) >DroSec_CAF1 8216 108 + 1 ----AGCAUGCAGCAGCAACUGCGAGG-UAAUCG-AAUGCGAUGUGGUCAAAAGGGCGUGACCGUUGGCAGGUGCACGUCAUAUUCGCACCUUUUGCGAACGACGCCCUUUC------CU ----.(((((((((((...))))((..-...)).-..))).))))((...(((((((((...((((.((((((((...........)))))...))).))))))))))))))------). ( -39.50) >DroEre_CAF1 7827 106 + 1 ----AGCAC---------ACUGCAAACUUAAUCAAAAUACGUUGUGGUCAAAAGGGCGUUAACA-GGCCAGGCGCAAGUCAUAUUUGCACCUUUUGCAUAUGACGCCCUUACGACGCUUU ----(((.(---------((.((.................)).)))(((..((((((((((...-.((.(((.((((((...)))))).)))...))...))))))))))..)))))).. ( -33.93) >DroYak_CAF1 7973 115 + 1 ACACAACAC----CGUCAACUGGGAACUUAAUCAAAAUGCGAUGUGGUCAAAAGGGCGUUAACA-GGCCAGGUGCAAGUCAUAUUUGCACCUUUUGCAAACGACGCCCUUGCGACGCUUU .((((.(.(----((.....)))((......)).......).))))(((..(((((((((....-.((.((((((((((...))))))))))...))....)))))))))..)))..... ( -38.00) >consensus ____AGCAC_____AGCAACUGCAAACUUAAUCAAAAUGCGAUGUGGUCAAAAGGGCGUGAACA_GGCCAGGUGCAAGUCAUAUUCGCACCUUUUGCAAACGACGCCCUUAC______CU ..................((((((..................))))))...(((((((((.........((((((((((...)))))))))).........))))))))).......... (-21.11 = -22.87 + 1.75)

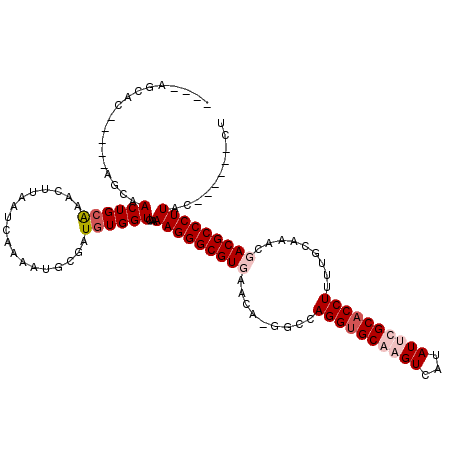

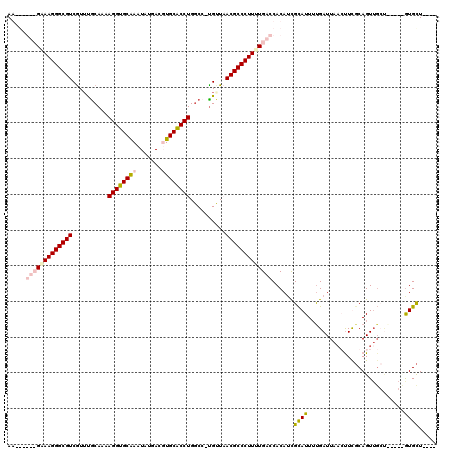
| Location | 17,442,987 – 17,443,093 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.67 |
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -26.24 |
| Energy contribution | -27.68 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17442987 106 - 22224390 AG------GGAAGGGCGUCGUUUACAAAAGGUGCGAAUAUGACGUGCACCUGGCC-UGUUCACGCCCUUUUGACCACAUUGCAUUUUGAUUAACUUUGCAGUUGCU---GUGUGCU---- ..------((((((((((.(...(((..(((((((.........)))))))....-))).)))))))))))...(((((.(((..(((..........))).))).---)))))..---- ( -33.40) >DroSec_CAF1 8216 108 - 1 AG------GAAAGGGCGUCGUUCGCAAAAGGUGCGAAUAUGACGUGCACCUGCCAACGGUCACGCCCUUUUGACCACAUCGCAUU-CGAUUA-CCUCGCAGUUGCUGCUGCAUGCU---- ..------((((((((((((((((((.....)))))..)))))(((.(((.......)))))))))))))).........((((.-.((...-..))(((((....))))))))).---- ( -39.50) >DroEre_CAF1 7827 106 - 1 AAAGCGUCGUAAGGGCGUCAUAUGCAAAAGGUGCAAAUAUGACUUGCGCCUGGCC-UGUUAACGCCCUUUUGACCACAACGUAUUUUGAUUAAGUUUGCAGU---------GUGCU---- ..(((((((.((((((((.(((.((...((((((((.......)))))))).)).-)))..)))))))).)))).(((..(((..((.....))..)))..)---------)))))---- ( -38.20) >DroYak_CAF1 7973 115 - 1 AAAGCGUCGCAAGGGCGUCGUUUGCAAAAGGUGCAAAUAUGACUUGCACCUGGCC-UGUUAACGCCCUUUUGACCACAUCGCAUUUUGAUUAAGUUCCCAGUUGACG----GUGUUGUGU .....((((.((((((((.....((...((((((((.......)))))))).)).-.....)))))))).))))((((.(((...(..(((........)))..)..----))).)))). ( -40.00) >consensus AA______GAAAGGGCGUCGUUUGCAAAAGGUGCAAAUAUGACGUGCACCUGGCC_UGUUAACGCCCUUUUGACCACAUCGCAUUUUGAUUAACUUCGCAGUUGCU_____GUGCU____ .....(((((((((((((..........((((((((.......))))))))..........)))))))))))))......((((...........................))))..... (-26.24 = -27.68 + 1.44)


| Location | 17,443,020 – 17,443,132 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -31.13 |
| Energy contribution | -33.63 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17443020 112 + 22224390 AAUGUGGUCAAAAGGGCGUGAACA-GGCCAGGUGCACGUCAUAUUCGCACCUUUUGUAAACGACGCCCUUCC------CUUAACAUUCGCCACAAAACCAGAA-GUCAGCUCUGUCAUAA ..((((((.(((((((((((.(((-((..((((((...........)))))))))))...).))))))))..------.......)).))))))....((((.-(....)))))...... ( -30.70) >DroSec_CAF1 8250 113 + 1 GAUGUGGUCAAAAGGGCGUGACCGUUGGCAGGUGCACGUCAUAUUCGCACCUUUUGCGAACGACGCCCUUUC------CUUACCGUUGGCCACAAAACCAGAA-GGCAUCGCUGUCAUAA ..(((((((((((((((((...((((.((((((((...........)))))...))).))))))))))))..------.......))))))))).........-((((....)))).... ( -42.30) >DroEre_CAF1 7854 119 + 1 GUUGUGGUCAAAAGGGCGUUAACA-GGCCAGGCGCAAGUCAUAUUUGCACCUUUUGCAUAUGACGCCCUUACGACGCUUUUUGCAUUGGCCACAAAACCAGUAAAGCAUCUCUGUCAUAA .((((((((((((((((((((...-.((.(((.((((((...)))))).)))...))...)))))))))).....((.....)).))))))))))...(((..........)))...... ( -42.10) >DroYak_CAF1 8009 119 + 1 GAUGUGGUCAAAAGGGCGUUAACA-GGCCAGGUGCAAGUCAUAUUUGCACCUUUUGCAAACGACGCCCUUGCGACGCUUUUUGCAUUGGCCACAAAACCAGAAGGGCAUCUCUGUCAUAA ..((((((((((((((((((....-.((.((((((((((...))))))))))...))....)))))))))((((......)))).)))))))))..........((((....)))).... ( -47.20) >consensus GAUGUGGUCAAAAGGGCGUGAACA_GGCCAGGUGCAAGUCAUAUUCGCACCUUUUGCAAACGACGCCCUUAC______CUUAGCAUUGGCCACAAAACCAGAA_GGCAUCUCUGUCAUAA ..((((((((((((((((((.........((((((((((...)))))))))).........)))))))))...............)))))))))....((((........))))...... (-31.13 = -33.63 + 2.50)

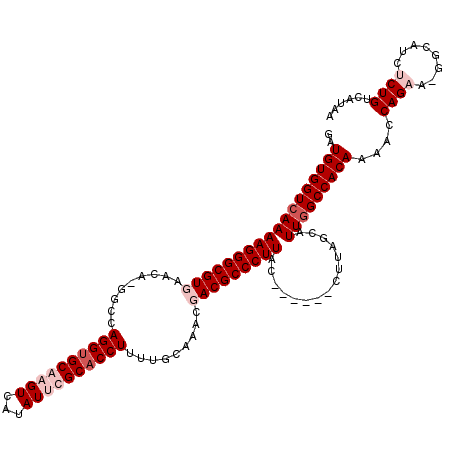
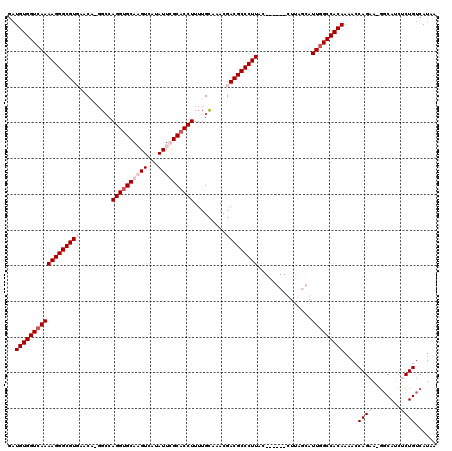
| Location | 17,443,020 – 17,443,132 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -39.15 |
| Consensus MFE | -31.01 |
| Energy contribution | -33.01 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17443020 112 - 22224390 UUAUGACAGAGCUGAC-UUCUGGUUUUGUGGCGAAUGUUAAG------GGAAGGGCGUCGUUUACAAAAGGUGCGAAUAUGACGUGCACCUGGCC-UGUUCACGCCCUUUUGACCACAUU ...(.((((((((...-....)))))))).)..(((((....------((((((((((.(...(((..(((((((.........)))))))....-))).)))))))))))....))))) ( -34.50) >DroSec_CAF1 8250 113 - 1 UUAUGACAGCGAUGCC-UUCUGGUUUUGUGGCCAACGGUAAG------GAAAGGGCGUCGUUCGCAAAAGGUGCGAAUAUGACGUGCACCUGCCAACGGUCACGCCCUUUUGACCACAUC ...((.(.((((.(((-....))).)))).).))..(((...------((((((((((((((((((.....)))))..)))))(((.(((.......)))))))))))))).)))..... ( -40.50) >DroEre_CAF1 7854 119 - 1 UUAUGACAGAGAUGCUUUACUGGUUUUGUGGCCAAUGCAAAAAGCGUCGUAAGGGCGUCAUAUGCAAAAGGUGCAAAUAUGACUUGCGCCUGGCC-UGUUAACGCCCUUUUGACCACAAC ......((((((((((((..(((((....)))))......))))))))...(((((((.(((.((...((((((((.......)))))))).)).-)))..)))))))))))........ ( -40.20) >DroYak_CAF1 8009 119 - 1 UUAUGACAGAGAUGCCCUUCUGGUUUUGUGGCCAAUGCAAAAAGCGUCGCAAGGGCGUCGUUUGCAAAAGGUGCAAAUAUGACUUGCACCUGGCC-UGUUAACGCCCUUUUGACCACAUC ...((((((.(((((((((((((((....)))))((((.....)))).).)))))))))....((...((((((((.......)))))))).)))-)))))................... ( -41.40) >consensus UUAUGACAGAGAUGCC_UUCUGGUUUUGUGGCCAAUGCAAAA______GAAAGGGCGUCGUUUGCAAAAGGUGCAAAUAUGACGUGCACCUGGCC_UGUUAACGCCCUUUUGACCACAUC ......(((((......)))))....(((((...............((((((((((((..........((((((((.......))))))))..........))))))))))))))))).. (-31.01 = -33.01 + 2.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:50 2006