| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,431,929 – 17,432,041 |
| Length | 112 |
| Max. P | 0.772713 |

| Location | 17,431,929 – 17,432,041 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -21.42 |
| Energy contribution | -20.73 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 17431929 112 + 22224390 UUUUUUU-UCCA--UUU--CCAGCGAACGUACUUCCUGGAUGCGGGUGCCAUUUCGGCGAUUUGCUUCACUGUCUUCGGCGUCUGCUGCACAGUGGGCACCAUUGGGAUCGUACUGU ......(-((((--..(--((((.(((.....))))))))....((((((.....(((.....))).((((((...((((....)))).))))))))))))..)))))......... ( -36.90) >DroVir_CAF1 30714 111 + 1 AUGG-UU-UUUG--AUU--AUAGCGCACCUAUUUUCUGGAUGCCGGCGCCAUCUCAGCCAUAUGCUUUACGGUCUUUGGUAUAUGCUGUACUGUGGGCACCAUUGGCAUUGUGCUGU ....-..-....--...--((((((((((........))(((((((.(((....((((.(((((((...........))))))))))).......))).))...))))))))))))) ( -34.50) >DroGri_CAF1 32858 112 + 1 AUUA-UG-UAUA-UGUU--AUAGCGCACAUAUUUUUUGGAUGCCGGCUCCAUAUCGGCCAUAUGUUUCACUGUCUUUGGCAUUUGCUGUACGGUGGGCACCAUUGGCAUCGUCCUUU ....-..-.(((-(((.--.......))))))......((((((((((.......))))...((((.((((((...((((....)))).)))))))))).....))))))....... ( -29.30) >DroWil_CAF1 16832 113 + 1 UUGU-GU-UUUA--UGCCCUUAGAGAACCUAUUUUCUGGAUGCUGGGGCCAUAUCAGCCAUUUGCUUUACAGUUUUUGGCAUUUGCUGCACAGUGGGCACCAUUGGCAUUGUCCUAU ....-..-....--.((..((((((((....))))))))..))((((((.......((((...(((....)))...))))......(((.((((((...)))))))))..)))))). ( -29.60) >DroMoj_CAF1 31330 111 + 1 UUUG-UU-UCUG--CUU--GCAGCGCACCUAUUUCCUGGACGCUGGCGCCAUCUCGGCCAUUUGCUUCACGGUCUUUGGCAUCUGCUGCACGGUGGGCACCAUUGGCAUUGUGCUAU ...(-(.-((..--(((--(((((((.((........))..))....((((....((((...........))))..))))....)))))).))..)).))...((((.....)))). ( -31.70) >DroAna_CAF1 11565 115 + 1 UCUAUCUUGUAAUCCUU--GUAGAGAACCUACUUCCUGGAUGCUGGUGCCAUUUCAGCGAUCUGCUUCACUGUAUUCGGUAUAUGCUGUACAGUGGGCACCAUUGGCAUCGUCCUGU .................--((((.....))))......((((((((((((.....(((.....))).(((((((..((((....)))))))))))))))))...))))))....... ( -35.10) >consensus UUUA_UU_UUUA__CUU__AUAGCGAACCUAUUUCCUGGAUGCUGGCGCCAUAUCAGCCAUUUGCUUCACUGUCUUUGGCAUAUGCUGCACAGUGGGCACCAUUGGCAUCGUCCUGU ......................................((((((((.(((.....(((.....))).((((((...((((....)))).))))))))).))...))))))....... (-21.42 = -20.73 + -0.69)


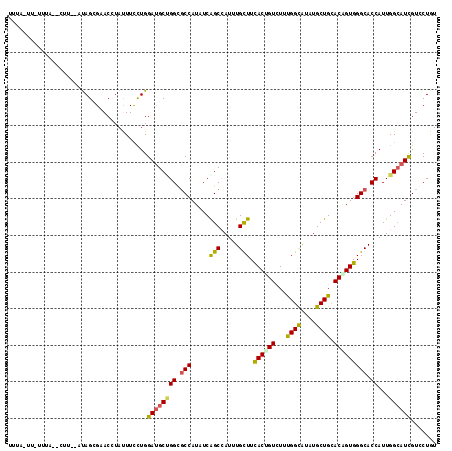
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:45 2006