
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 17,424,255 – 17,424,371 |
| Length | 116 |
| Max. P | 0.919748 |

| Location | 17,424,255 – 17,424,371 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.91 |
| Mean single sequence MFE | -39.38 |
| Consensus MFE | -22.28 |
| Energy contribution | -23.96 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
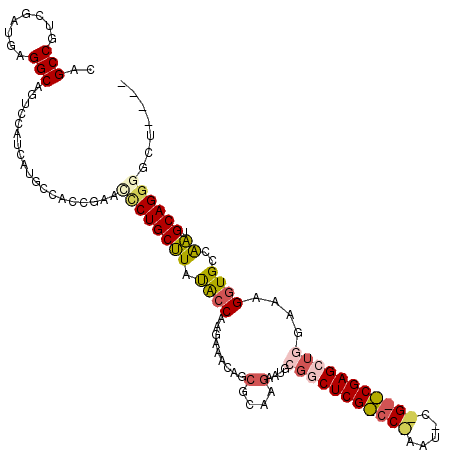
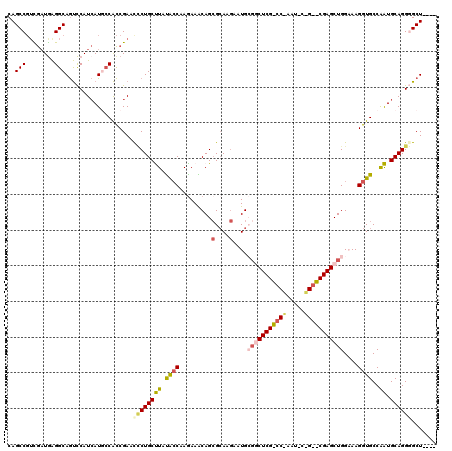
>X_DroMel_CAF1 17424255 116 + 22224390 CAGCCGUCGAUGAGGCAGUCCAUCAUUCCACCGAAUCCUGCUUAUAGCAAGAAACAGGGCAAAAAUGCGGCUCGCCCAAAUUCCGAGCGAGCUGGAAAGGUGCCAAUGCAGGGGCU---- ..(((........)))(((((..((((........((.(((.....))).)).....((((....(.((((((((.(.......).)))))))).)....))))))))...)))))---- ( -37.50) >DroSec_CAF1 348 95 + 1 CAGCCGUCGAUGAGGCAGUCCAUCAUGCCACCGAACCCUGCCUAUACCAAGAAACAGCGCAAGAAUGCGC---------------------CUGGAAAGGUGCCAGUGCAGGGGCU---- .((((.(((....((((........))))..)))..(((((((..............(....)...((((---------------------((....)))))).)).)))))))))---- ( -34.00) >DroSim_CAF1 332 95 + 1 CAGCCGUCGAUGAGGCAGUCCAUCAUGCCACCGAACCCUGCCUAUACCAAGAAACAGCGCAAGAAUGCGG---------------------CUGGAAAGGUGCCAGUGCAGGGGCU---- .((((.(((....((((........))))..)))..(((((((.((((......(((((((....))).)---------------------)))....))))..)).)))))))))---- ( -33.70) >DroEre_CAF1 94 119 + 1 CAGCCGUUGAUAGGGCAGCUCAUCAUAACGCCGAACGCUGCUUACGCCAAGAAGCAGCGGAAGACUGGGGCUCGUCCAAAUCCUGGACGAGCUGGCAAGGUGCCGAUGCAGGUGCUGGU- ..(((((((......))))..........(((...((((((((.(.....))))))))).........((((((((((.....)))))))))))))..((..((......))..)))))- ( -49.60) >DroYak_CAF1 358 120 + 1 CAGCCUAUAUUGAGGCAAUCCAUAAUGCCAUCGAACUCUGCUUAUGCCAAAAAGCAGCGCAAGACUGGCACUCGUCCGAAUCCUGGACGAGCUGGUAAGGUGCCGAUGCAGGUGCUCGUU ..((((......))))...(((...(((((((...(.((((((........)))))).)...)).)))))((((((((.....)))))))).)))...((..((......))..)).... ( -42.10) >consensus CAGCCGUCGAUGAGGCAGUCCAUCAUGCCACCGAACCCUGCUUAUACCAAGAAACAGCGCAAGAAUGCGGCUCG_CC_AAU_C_G__CGAGCUGGAAAGGUGCCAAUGCAGGGGCU____ ..(((........)))...................((((((((.((((.........(....)....(((((((((((.....)))))))))))....))))..)).))))))....... (-22.28 = -23.96 + 1.68)

| Location | 17,424,255 – 17,424,371 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.91 |
| Mean single sequence MFE | -41.60 |
| Consensus MFE | -24.16 |
| Energy contribution | -25.38 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
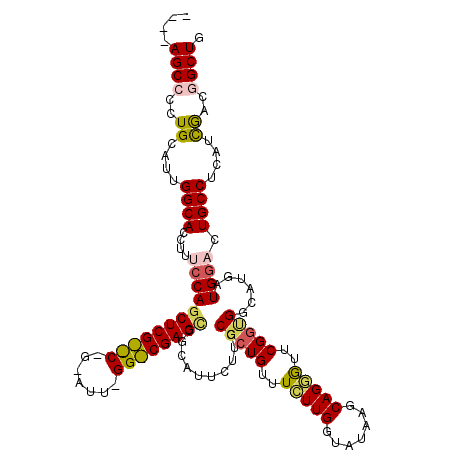
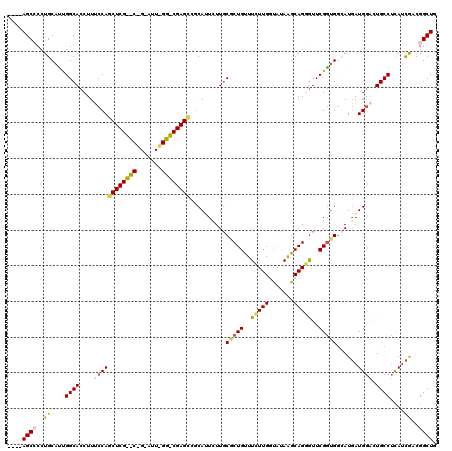
>X_DroMel_CAF1 17424255 116 - 22224390 ----AGCCCCUGCAUUGGCACCUUUCCAGCUCGCUCGGAAUUUGGGCGAGCCGCAUUUUUGCCCUGUUUCUUGCUAUAAGCAGGAUUCGGUGGAAUGAUGGACUGCCUCAUCGACGGCUG ----((((..((....((((((.((((((((((((((.....))))))))).(((....))).(((..((((((.....))))))..))))))))....))..))))....))..)))). ( -44.40) >DroSec_CAF1 348 95 - 1 ----AGCCCCUGCACUGGCACCUUUCCAG---------------------GCGCAUUCUUGCGCUGUUUCUUGGUAUAGGCAGGGUUCGGUGGCAUGAUGGACUGCCUCAUCGACGGCUG ----(((((((((.(((..(((......(---------------------(((((....)))))).......))).))))))))..(((((((((........))))..))))).)))). ( -38.82) >DroSim_CAF1 332 95 - 1 ----AGCCCCUGCACUGGCACCUUUCCAG---------------------CCGCAUUCUUGCGCUGUUUCUUGGUAUAGGCAGGGUUCGGUGGCAUGAUGGACUGCCUCAUCGACGGCUG ----(((((((((.(((..(((....(((---------------------(.(((....)))))))......))).))))))))..(((((((((........))))..))))).)))). ( -36.50) >DroEre_CAF1 94 119 - 1 -ACCAGCACCUGCAUCGGCACCUUGCCAGCUCGUCCAGGAUUUGGACGAGCCCCAGUCUUCCGCUGCUUCUUGGCGUAAGCAGCGUUCGGCGUUAUGAUGAGCUGCCCUAUCAACGGCUG -..((((...((....((((.(((..((((((((((((...))))))))))....(((...((((((((........))))))))...)))....))..))).))))....))...)))) ( -45.40) >DroYak_CAF1 358 120 - 1 AACGAGCACCUGCAUCGGCACCUUACCAGCUCGUCCAGGAUUCGGACGAGUGCCAGUCUUGCGCUGCUUUUUGGCAUAAGCAGAGUUCGAUGGCAUUAUGGAUUGCCUCAAUAUAGGCUG ..(((((....)).)))........(((((((((((.......))))))))((((.((..((.((((((........)))))).))..))))))....)))...((((......)))).. ( -42.90) >consensus ____AGCCCCUGCAUUGGCACCUUUCCAGCUCG__C_G_AUU_GG_CGAGCCGCAUUCUUGCGCUGUUUCUUGGUAUAAGCAGGGUUCGGUGGCAUGAUGGACUGCCUCAUCGACGGCUG ....((((..((....((((....((((((((((((.......))))))))..........(((((..(((((.......)))))..)))))......)))).))))....))..)))). (-24.16 = -25.38 + 1.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:26:44 2006